7Q1R
 
 | | A de novo designed homo-dimeric antiparallel coiled coil apCC-Di | | Descriptor: | ETHANOL, SODIUM ION, apCC-Di | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
6PBE
 
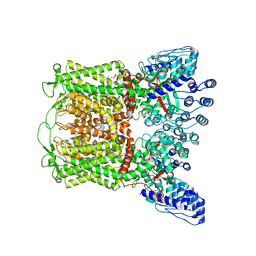 | | ZINC17988990-bound TRPV5 in nanodiscs | | Descriptor: | (4-oxo-5-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-2-yl)methyl 3-(3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)propanoate, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Rosario, J.S.D, Kapoor, A, Yazici, A.T, Fluck, E.C, Filizola, M, Rohacs, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure-based characterization of novel TRPV5 inhibitors.
Elife, 8, 2019
|
|
7PZT
 
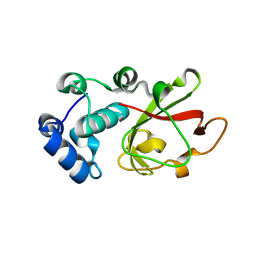 | | Structure of the bacterial toxin, TecA, an asparagine deamidase from Alcaligenes faecalis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Urea amidohydrolase | | Authors: | Dix, S.R, Aziz, A.A, Baker, P.J, Evans, C.A, Dickman, M.J, Farthing, R.J, King, Z.L.S, Nathan, S, Partridge, L.J, Raih, F.M, Sedelnikova, S.E, Thomas, M.S, Rice, D.W. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of A. faecalis TecA provides insights into its role as an asparagine deamidase toxin which targets RhoA
To Be Published
|
|
6UVV
 
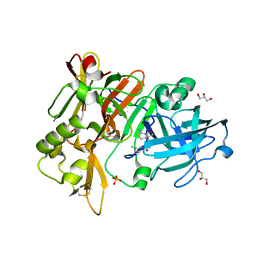 | | BACE-1 in complex with compound #17 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-butylcyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6YA6
 
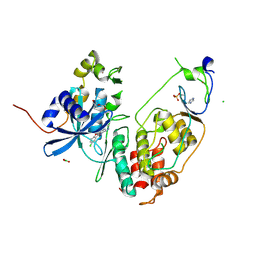 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6F6Y
 
 | |
6UXZ
 
 | | (S)-4-Amino-5-phenoxypentanoate as a Selective Agonist of the Transcription Factor GabR | | Descriptor: | (4S)-4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-5-phenoxypentanoic acid, HTH-type transcriptional regulatory protein GabR, SULFATE ION | | Authors: | Catlin, D.S, Liu, D. | | Deposit date: | 2019-11-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | (S)-4-Amino-5-phenoxypentanoate designed as a potential selective agonist of the bacterial transcription factor GabR.
Protein Sci., 29, 2020
|
|
5KBD
 
 | |
6Y48
 
 | |
6MRK
 
 | |
8EYU
 
 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8C31
 
 | |
6N3A
 
 | | SegA-long, conformation of TDP-43 low complexity domain segment A long | | Descriptor: | TAR DNA-binding protein 43, segA long small | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4XX1
 
 | | Low resolution structure of LCAT in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 heavy chain, Fab1 light chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
8EYV
 
 | | Structure of Beetroot dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (45-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8F0N
 
 | | Wobble Beetroot (A16U-U38G) dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-11-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
6N8Y
 
 | | Hsp90-beta bound to PU-11-trans | | Descriptor: | 9-[(2E)-but-2-en-1-yl]-8-[(3,4,5-trimethoxyphenyl)methyl]-9H-purin-6-amine, Heat shock protein HSP 90-beta | | Authors: | Huck, J.D, Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2018-11-30 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.553908 Å) | | Cite: | Structures of Hsp90 alpha and Hsp90 beta bound to a purine-scaffold inhibitor reveal an exploitable residue for drug selectivity.
Proteins, 87, 2019
|
|
8EFY
 
 | |
6MJX
 
 | | human cGAS catalytic domain bound with cGAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
8EFV
 
 | |
8KFV
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
6EYT
 
 | |
6N9J
 
 | |
5JWE
 
 | | Crystal structure of H-2Db in complex with the LCMV-derived GP92-101 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
8K88
 
 | | Structure of procaryotic Ago | | Descriptor: | DNA (41-mer), DNA/RNA (21-mer), MAGNESIUM ION, ... | | Authors: | Gao, X, Sun, D, Cui, S, Wang, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of prokaryotic Argonaute System
To Be Published
|
|
