7MVU
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 1-1756; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
6E5W
 
 | | Crystal structure of human cellular retinol binding protein 3 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, GLYCEROL, Retinol-binding protein 5 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
8G4W
 
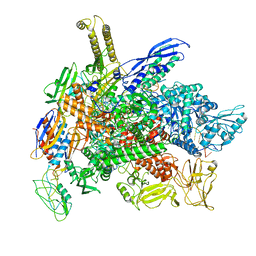 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7RMN
 
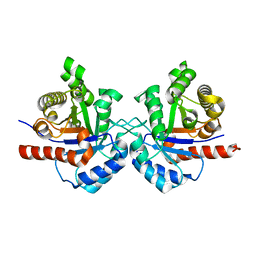 | |
8G8Z
 
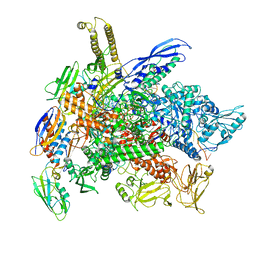 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | Authors: | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5L3J
 
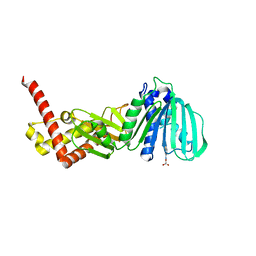 | | ESCHERICHIA COLI DNA GYRASE B IN COMPLEX WITH BENZOTHIAZOLE-BASED INHIBITOR | | Descriptor: | 2-[[2-[[4,5-bis(bromanyl)-1~{H}-pyrrol-2-yl]carbonylamino]-1,3-benzothiazol-6-yl]amino]-2-oxidanylidene-ethanoic acid, DNA gyrase subunit B, IODIDE ION | | Authors: | Gjorgjieva, M, Tomasic, T, Barancokova, M, Katsamakas, S, Ilas, J, Montalvao, S, Tammella, P, Peterlin Masic, L, Kikelj, D. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Benzothiazole Scaffold-Based DNA Gyrase B Inhibitors.
J.Med.Chem., 59, 2016
|
|
7MVY
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
3GMD
 
 | | Structure-based design of 7-Azaindole-pyrrolidines as inhibitors of 11beta-Hydroxysteroid-Dehydrogenase type I | | Descriptor: | 2-methyl-3-{(3S)-1-[(1-pyridin-2-ylcyclopropyl)carbonyl]pyrrolidin-3-yl}-1H-pyrrolo[2,3-b]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Valeur, E, Lepifre, F, Roche, D, Christmann-Franck, S, Hillertz, P, Musil, D. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11 beta-hydroxysteroid dehydrogenase type I.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7MW0
 
 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | Descriptor: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5L1U
 
 | |
4WXQ
 
 | | Crystal Structure of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Myocilin, ... | | Authors: | Orwig, S.D, Turnage, K.C, Donegan, R.K, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
5XG5
 
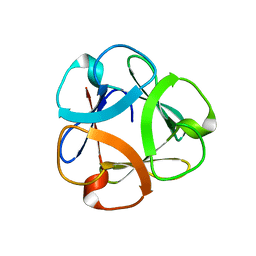 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
6ZLX
 
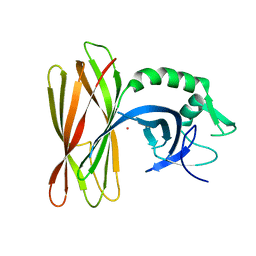 | | The structure of the ClpX-associated factor PDIP38 | | Descriptor: | PLATINUM (II) ION, Polymerase delta-interacting protein 2 | | Authors: | Zeth, K, Dougan, D. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | PDIP38 is a novel adaptor-like modulator of the mitochondrial AAA+ protease CLPXP
To Be Published
|
|
6EI9
 
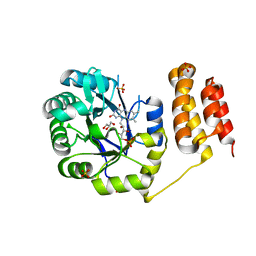 | | Crystal structure of E. coli tRNA-dihydrouridine synthase B (DusB) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Hamdane, D. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario.
Nucleic Acids Res., 46, 2018
|
|
7OYB
 
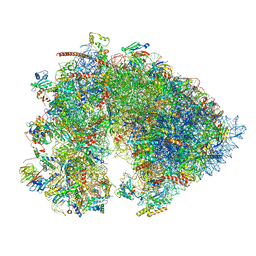 | | Cryo-EM structure of the 6 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7R6P
 
 | |
6ZOI
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conknunitzin-C3 mutante | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A lid blocking mechanism of a cone snail toxin revealed at the atomic level
To Be Published
|
|
7OYA
 
 | | Cryo-EM structure of the 1 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
6QUH
 
 | | GHK tagged GFP variant crystal form II at 1.34A wavelength | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5L8Z
 
 | | Structure of thermostable DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein, SODIUM ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korzhenevskiy, D.A, Kamashev, D.E, Vanyushkina, A.A, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the high thermal stability of the histone-like HU protein from the mollicute Spiroplasma melliferum KC3.
Sci Rep, 6, 2016
|
|
8SZE
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim
To be published
|
|
6ZWK
 
 | | Crystal structure of the phosphorylated C-terminal tail of histone H2AX in complex with a specific nanobody (C6 gammaXbody) | | Descriptor: | CHLORIDE ION, Histone H2AX, SODIUM ION, ... | | Authors: | McEwen, A.G, Moeglin, E, Desplancq, D, Weiss, E, Poterszman, A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Novel Nanobody Precisely Visualizes Phosphorylated Histone H2AX in Living Cancer Cells under Drug-Induced Replication Stress.
Cancers (Basel), 13, 2021
|
|
6ED0
 
 | |
8SZD
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, MAGNESIUM ION, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution
To be published
|
|
8G7E
 
 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
