2WA9
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - Trp peptide structure | | 分子名称: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, TRP PEPTIDE | | 著者 | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | 登録日 | 2009-02-03 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
4W6I
 
 | |
4RCF
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 4-fluoroxanthene inhibitor 49 | | 分子名称: | (4S)-2'-(3,6-dihydro-2H-pyran-4-yl)-4'-fluoro-7'-(2-fluoropyridin-3-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, GLYCEROL, ... | | 著者 | Whittington, D.A, Long, A.M. | | 登録日 | 2014-09-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Lead Optimization and Modulation of hERG Activity in a Series of Aminooxazoline Xanthene beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4W77
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 3 | | 分子名称: | COPPER (II) ION, fluorescent protein D21H/K26C | | 著者 | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | 登録日 | 2014-08-21 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
3E4D
 
 | | Structural and Kinetic Study of an S-Formylglutathione Hydrolase from Agrobacterium tumefaciens | | 分子名称: | CHLORIDE ION, Esterase D, MAGNESIUM ION | | 著者 | Van Straaten, K.E, Gonzalez, C.F, Valladares, R.B, Xu, X, Savchenko, A.V, Sanders, D.A.R. | | 登録日 | 2008-08-11 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens
Protein Sci., 18, 2009
|
|
7AMP
 
 | | Crystal structure of the complex of HuJovi-1 Fab with the human A6 T-cell receptor TRBC1 | | 分子名称: | Alpha chain of A6 T-cell receptor, Beta chain 1 of A6 T-cell receptor TRBC1, CHLORIDE ION, ... | | 著者 | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | 登録日 | 2020-10-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
7AMQ
 
 | | Crystal structure of the complex of HuJovi-1 Fab with the human TRBC2 | | 分子名称: | CHLORIDE ION, GLYCEROL, Human A6 T-cell receptor alpha chain, ... | | 著者 | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | 登録日 | 2020-10-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-04-27 | | 実験手法 | X-RAY DIFFRACTION (2.353 Å) | | 主引用文献 | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
7AMS
 
 | | Crystal structure of the complex of the KFN mutant of HuJovi-1 Fab with human TRBC2 | | 分子名称: | CHLORIDE ION, GLYCEROL, Human A6 T-cell receptor alpha chain, ... | | 著者 | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Svensson, A, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | 登録日 | 2020-10-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-04-27 | | 実験手法 | X-RAY DIFFRACTION (2.419 Å) | | 主引用文献 | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
4V7J
 
 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | 登録日 | 2009-11-02 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
7AMR
 
 | | Crystal structure of the complex of the KFN mutant of Jovi-1 Fab with human TRBC1 | | 分子名称: | CHLORIDE ION, GLYCEROL, Human Jovi-1 Fab fragment, ... | | 著者 | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | 登録日 | 2020-10-09 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-04-27 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
5DF1
 
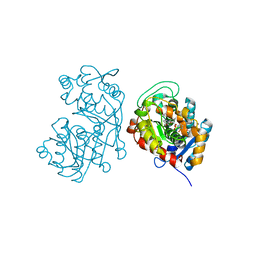 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and geranic acid | | 分子名称: | (2E)-3,7-dimethylocta-2,6-dienoic acid, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | 著者 | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | 登録日 | 2015-08-26 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
1JYJ
 
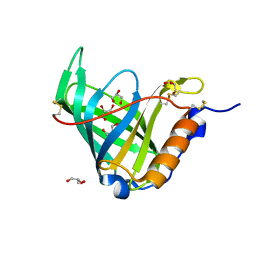 | | Crystal Structure of a Double Variant (W67L/W91H) of Recombinant Human Serum Retinol-binding Protein at 2.0 A Resolution | | 分子名称: | GLYCEROL, PLASMA RETINOL-BINDING PROTEIN | | 著者 | Greene, L.H, Chrysina, E.D, Irons, L.I, Papageorgiou, A.C, Acharya, K.R, Brew, K. | | 登録日 | 2001-09-12 | | 公開日 | 2003-07-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Role of Conserved Residues in Structure and Stability: Tryptophans of Human Serum Retinol-Binding Protein, a Model for the Lipocalin Superfamily
Protein Sci., 10, 2001
|
|
8I40
 
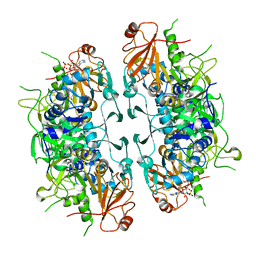 | | Crystal structure of ASCT from Trypanosoma brucei in complex with CoA. | | 分子名称: | ACETATE ION, CALCIUM ION, COENZYME A, ... | | 著者 | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | 登録日 | 2023-01-18 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structure of ligand complexes of ASCT from Trypanosoma brucei and molecular mechanism in comparison with mammalian SCOT.
To Be Published
|
|
6IZH
 
 | |
2IK8
 
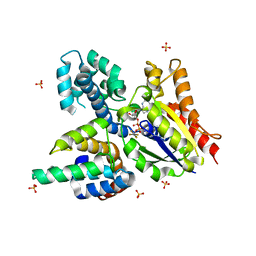 | | Crystal structure of the heterodimeric complex of human RGS16 and activated Gi alpha 1 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | 著者 | Soundararajan, M, Turnbull, A.P, Papagrigoriou, E, Debreczeni, J, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | 登録日 | 2006-10-02 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5D12
 
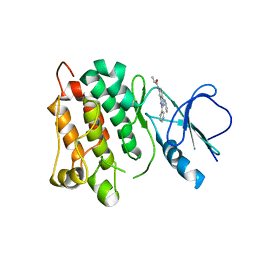 | | Kinase domain of cSrc in complex with RL40 | | 分子名称: | N-[2-phenyl-4-(1H-pyrazol-3-ylamino)quinazolin-7-yl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Becker, C, Richters, A, Engel, J, Rauh, D. | | 登録日 | 2015-08-03 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
5KK0
 
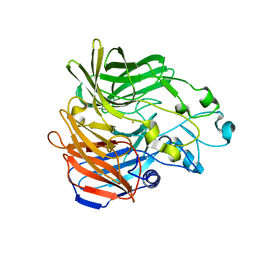 | | Synechocystis ACO mutant - T136A | | 分子名称: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | 著者 | Sui, X, Kiser, P.D, Palczewski, K. | | 登録日 | 2016-06-20 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Utilization of Dioxygen by Carotenoid Cleavage Oxygenases.
J.Biol.Chem., 290, 2015
|
|
7ZUV
 
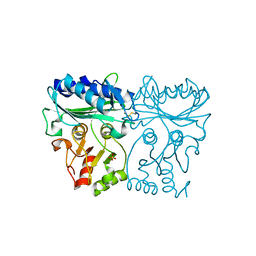 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic sedoheptulose-1,7-bisphosphatase in reducing conditions | | 分子名称: | FBPase domain-containing protein, SULFATE ION | | 著者 | Le Moigne, T, Robert, G.Q, Lemaire, S.D, Henri, J. | | 登録日 | 2022-05-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Characterization of chloroplast ribulose-5-phosphate-3-epimerase from the microalga Chlamydomonas reinhardtii.
Plant Physiol., 194, 2024
|
|
7ZQ7
 
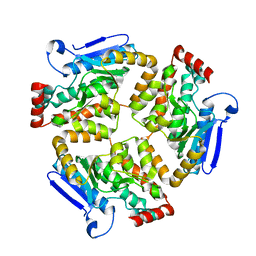 | | Structure of RpF-1 | | 分子名称: | Crotonase/enoyl-CoA hydratase family protein | | 著者 | Sanchez-Alba, L, Reverter, D, Conchillo, O, Yero, D, Daura, X, Gibert, I. | | 登録日 | 2022-04-29 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of RpF-1
To Be Published
|
|
2W5P
 
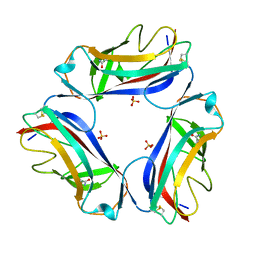 | | DraE Adhesin in complex with Chloramphenicol Succinate (monoclinic form) | | 分子名称: | CHLORAMPHENICOL SUCCINATE, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION | | 著者 | Pettigrew, D.M, Roversi, P, Davies, S.G, Russell, A.J, Lea, S.M. | | 登録日 | 2008-12-11 | | 公開日 | 2009-06-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Structural Study of the Interaction between the Dr Haemagglutinin Drae and Derivatives of Chloramphenicol
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6W7L
 
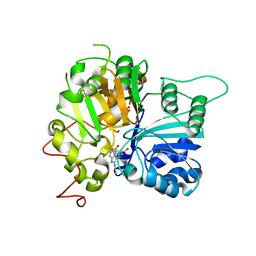 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ632p | | 分子名称: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyrazin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | 著者 | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.856 Å) | | 主引用文献 | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
6W7K
 
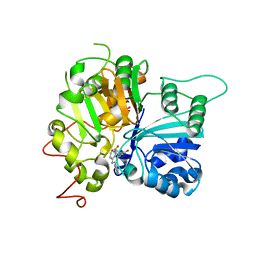 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ634p | | 分子名称: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyridin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | 著者 | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
5D72
 
 | | Crystal structure of MOR04252, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | 分子名称: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | 著者 | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | 登録日 | 2015-08-13 | | 公開日 | 2015-10-14 | | 最終更新日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5KRL
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the A-CD ring estrogen, (1S,7aS)-5-(2-chloro-4-hydroxyphenyl)-7a-methyl-2,3,3a,4,7,7a-hexahydro-1H-inden-1-ol | | 分子名称: | (1~{S},3~{a}~{R},7~{a}~{S})-5-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,7,7~{a}-hexahydro-1~{H}-inden-1-ol, Estrogen receptor, NCOA2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-07-07 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2W5V
 
 | | Structure of TAB5 alkaline phosphatase mutant His 135 Asp with Mg bound in the M3 site. | | 分子名称: | ALKALINE PHOSPHATASE, MAGNESIUM ION, ZINC ION | | 著者 | Koutsioulis, D, Lyskowski, A, Maki, S, Guthrie, E, Feller, G, Bouriotis, V, Heikinheimo, P. | | 登録日 | 2008-12-15 | | 公開日 | 2009-11-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Coordination Sphere of the Third Metal Site is Essential to the Activity and Metal Selectivity of Alkaline Phosphatases.
Protein Sci., 19, 2010
|
|
