8CX2
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | 分子名称: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | 著者 | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | 登録日 | 2022-05-19 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
6G5T
 
 | | Myoglobin H64V/V68A in the resting state, 1.5 Angstrom resolution | | 分子名称: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | 登録日 | 2018-03-30 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
7KBA
 
 | |
8I5E
 
 | | Crystal structure of HLA-A*11:01 in complex with KRAS peptide (VVGAGGVGK) | | 分子名称: | Beta-2-microglobulin, KRAS-G12wt-9, MHC class I antigen (Fragment) | | 著者 | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | 登録日 | 2023-01-25 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
5LPM
 
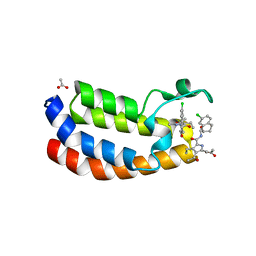 | | Crystal structure of the bromodomain of human Ep300 bound to the inhibitor XDM3d | | 分子名称: | ACETATE ION, Histone acetyltransferase p300, ~{N}-[(1~{S},2~{S})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole -2-carboxamide | | 著者 | Huegle, M, Wohlwend, D, Gerhardt, S. | | 登録日 | 2016-08-13 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5KCT
 
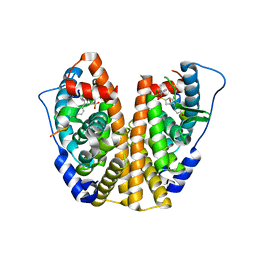 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | 分子名称: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-06-07 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
8CX1
 
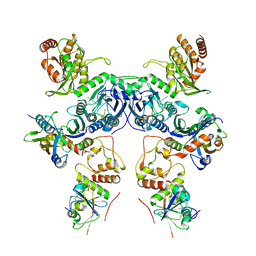 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | 分子名称: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | 著者 | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | 登録日 | 2022-05-19 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
7Q1Q
 
 | | De novo designed homo-dimeric antiparallel helices Homomer-S | | 分子名称: | ACETATE ION, Homomer-S | | 著者 | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | 登録日 | 2021-10-20 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
6QT9
 
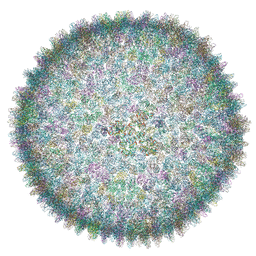 | | Cryo-EM structure of SH1 full particle. | | 分子名称: | ORF 24, ORF 25, ORF 31, ... | | 著者 | De Colibus, L, Roine, E, Walter, T.S, Ilca, S.L, Wang, X, Wang, N, Roseman, A.M, Bamford, D, Huiskonen, J.T, Stuart, D.I. | | 登録日 | 2019-02-22 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Assembly of complex viruses exemplified by a halophilic euryarchaeal virus.
Nat Commun, 10, 2019
|
|
4WES
 
 | | Nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 1.08 A resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, ... | | 著者 | Zhang, L.M, Morrison, C.N, Kaiser, J.T, Rees, D.C. | | 登録日 | 2014-09-10 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 angstrom resolution: comparison with the Azotobacter vinelandii MoFe protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7B5G
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | 分子名称: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | 著者 | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | 登録日 | 2020-12-03 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
5TQZ
 
 | | Frutapin complexed with alpha-D-glucose | | 分子名称: | Frutapin, alpha-D-glucopyranose | | 著者 | Sousa, F.D, Guo, J, Coker, A.R, Monteiro-Moreira, A.C.O, Moreira, R.A. | | 登録日 | 2016-10-25 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Frutapin, a lectin fromArtocarpus incisa(breadfruit): cloning, expression and molecular insights.
Biosci. Rep., 37, 2017
|
|
6H1X
 
 | |
3G9W
 
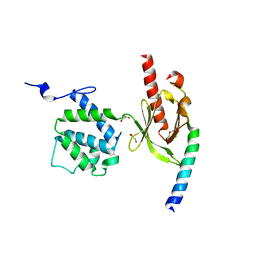 | | Crystal Structure of Talin2 F2-F3 in Complex with the Integrin Beta1D Cytoplasmic Tail | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Integrin beta-1D, ... | | 著者 | Anthis, N.J, Wegener, K.L, Ye, F, Kim, C, Lowe, E.D, Vakonakis, I, Bate, N, Critchley, D.R, Ginsberg, M.H, Campbell, I.D. | | 登録日 | 2009-02-15 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.165 Å) | | 主引用文献 | The structure of an integrin/talin complex reveals the basis of inside-out signal transduction
Embo J., 28, 2009
|
|
8PMQ
 
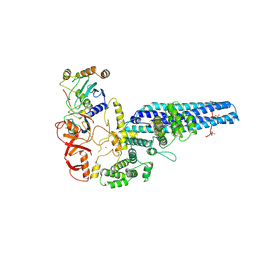 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | 分子名称: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | 登録日 | 2023-06-29 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
6W2E
 
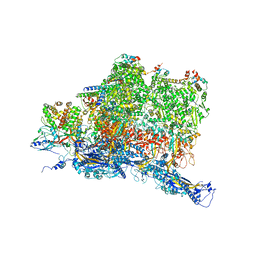 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | 登録日 | 2020-03-05 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
8HZZ
 
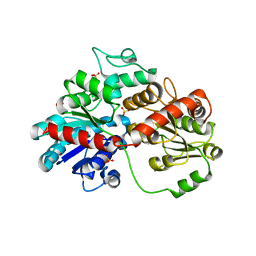 | | GuApiGT (UGT79B74) | | 分子名称: | SULFATE ION, apiosyltransferase | | 著者 | Wang, H.T, Wang, Z.L, Li, F.D, Ye, M. | | 登録日 | 2023-01-10 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
3G4F
 
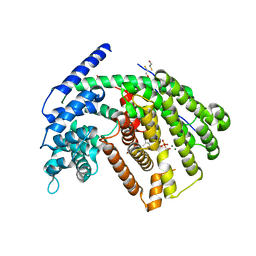 | | Crystal Structure of (+)- -Cadinene Synthase from Gossypium arboreum in complex with 2-fluorofarnesyl diphosphate | | 分子名称: | (+)-delta-cadinene synthase isozyme XC1, (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, BETA-MERCAPTOETHANOL, ... | | 著者 | Gennadios, H.A, Di Costanzo, L, Christianson, D.W. | | 登録日 | 2009-02-03 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.651 Å) | | 主引用文献 | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
5N6M
 
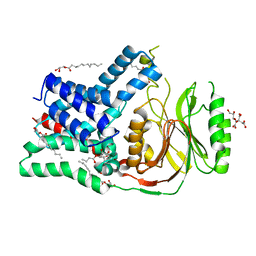 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | 著者 | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | 登録日 | 2017-02-15 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
7A97
 
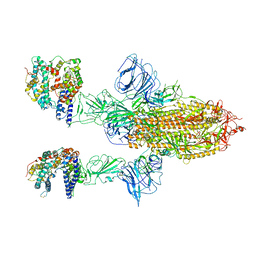 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | 登録日 | 2020-09-01 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
6GUZ
 
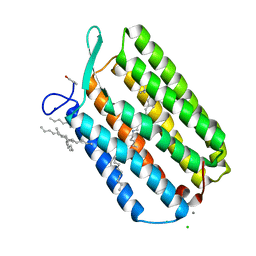 | | Ground state structure of Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich at room temperature | | 分子名称: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Moraes, I, Judge, P.J, Bada Juarez, J.F, Vinals, J, Axford, D, Watts, A. | | 登録日 | 2018-06-19 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Room temperature structure of the Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich
To Be Published
|
|
8UHB
 
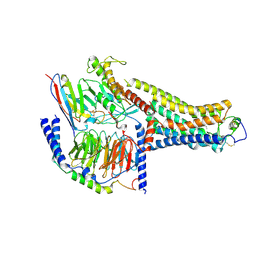 | | Cryo-EM Structure of the Ro5256390-bound hTA1-Gs heterotrimer signaling complex | | 分子名称: | (2R,4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zilberg, G, Warren, A.L, Parpounas, A.K, Wacker, D. | | 登録日 | 2023-10-08 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Molecular basis of human trace amine-associated receptor 1 activation.
Nat Commun, 15, 2024
|
|
3GDG
 
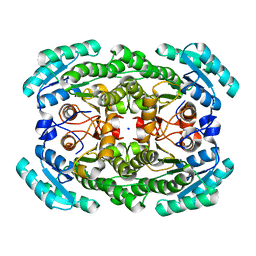 | | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum. | | 分子名称: | Probable NADP-dependent mannitol dehydrogenase, SODIUM ION | | 著者 | Nuess, D, Goettig, P, Magler, I, Denk, U, Breitenbach, M, Schneider, P.B, Brandstetter, H, Simon-Nobbe, B. | | 登録日 | 2009-02-24 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
Biochimie, 92, 2010
|
|
5H37
 
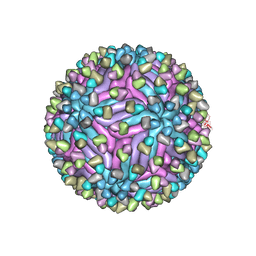 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 8.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C10 IgG heavy chain variable region, C10 IgG light chain variable region, ... | | 著者 | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lim, X.-N, Ooi, J.S.G, Lambert, S, Tan, T.Y, Widman, D, Shi, J, Baric, R.S, Lok, S.-M. | | 登録日 | 2016-10-20 | | 公開日 | 2016-11-30 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
7S23
 
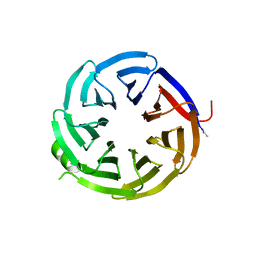 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | 分子名称: | Coatomer subunit alpha | | 著者 | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | 登録日 | 2021-09-03 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
