5K4B
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | 分子名称: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
8P2C
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric state produced in the presence of dATP and CTP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | 著者 | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | 登録日 | 2023-05-15 | | 公開日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
4YHS
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Monosaccharide ABC transporter substrate-binding protein, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2015-02-27 | | 公開日 | 2015-03-18 | | 最終更新日 | 2015-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS
To be published
|
|
5FQL
 
 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | 登録日 | 2015-12-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
4YI9
 
 | |
5MUU
 
 | | dsRNA bacteriophage phi6 nucleocapsid | | 分子名称: | Major inner protein P1, Major outer capsid protein, Packaging enzyme P4 | | 著者 | Sun, Z, El Omari, K, Sun, X, Ilca, S.L, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | 登録日 | 2017-01-14 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
6XB2
 
 | | Room temperature X-ray crystallography reveals catalytic cysteine in the SARS-CoV-2 3CL Mpro is highly reactive: Insights for enzyme mechanism and drug design | | 分子名称: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Kneller, D.W, Kovalevsky, A, Coates, L. | | 登録日 | 2020-06-05 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Room-temperature X-ray crystallography reveals the oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL M pro : insights into enzyme mechanism and drug design.
Iucrj, 7, 2020
|
|
6I53
 
 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | 著者 | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-02 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
8ETQ
 
 | |
6BT5
 
 | | Human mGlu8 Receptor complexed with L-AP4 | | 分子名称: | (2S)-2-amino-4-phosphonobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 8 | | 著者 | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | 登録日 | 2017-12-05 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6N6F
 
 | |
6FEL
 
 | |
6XTK
 
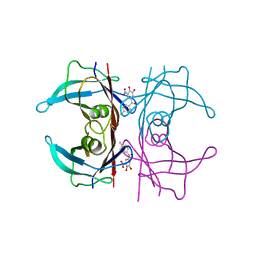 | | Y114C Transthyretin structure in complex with Tolcalpone | | 分子名称: | Tolcapone, Transthyretin | | 著者 | Varejao, N, Reverter, D, Pinheiro, F, Pallares, I, Ventura, S. | | 登録日 | 2020-01-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis.
Febs J., 288, 2021
|
|
5UFC
 
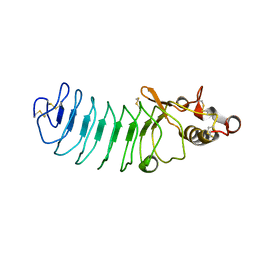 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 with H-trisaccharide bound | | 分子名称: | Tn4-22, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | 登録日 | 2017-01-04 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.888 Å) | | 主引用文献 | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5KB3
 
 | |
6V1Y
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Octamer | | 分子名称: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | 著者 | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | 登録日 | 2019-11-21 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
6FJS
 
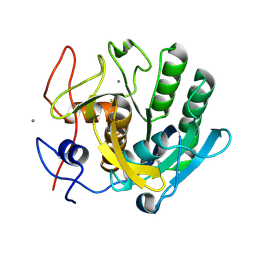 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | 分子名称: | CALCIUM ION, Proteinase K | | 著者 | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
4Y7I
 
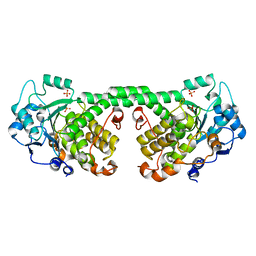 | | Crystal Structure of MTMR8 | | 分子名称: | Myotubularin-related protein 8, PHOSPHATE ION | | 著者 | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | 登録日 | 2015-02-15 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6BSZ
 
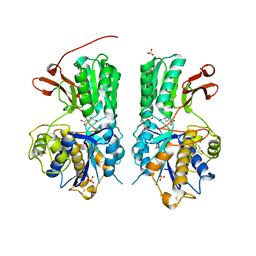 | | Human mGlu8 Receptor complexed with glutamate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 8, ... | | 著者 | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | 登録日 | 2017-12-04 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5CDR
 
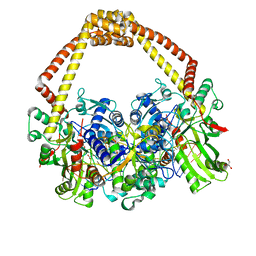 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Bax, B.D, Srikannathasan, V, Chan, P.F. | | 登録日 | 2015-07-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
4YFT
 
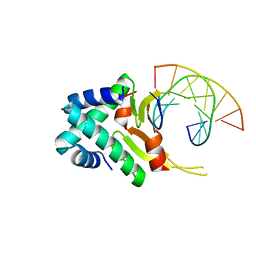 | | HUab-20bp | | 分子名称: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | 著者 | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | 登録日 | 2015-02-25 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.914 Å) | | 主引用文献 | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
5JQX
 
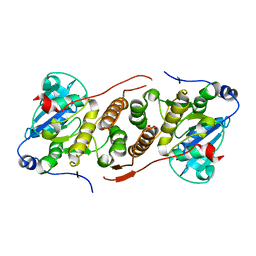 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with phosphoglyceric acid (PGA) - GpgS*PGA | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase | | 著者 | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | 登録日 | 2016-05-05 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
7YSF
 
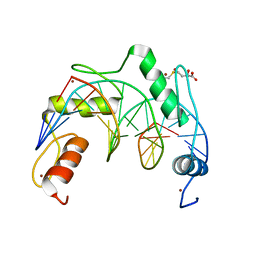 | | Crystal structure of ZNF524 ZF1-4 in complex with telomeric DNA | | 分子名称: | D-MALATE, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*C)-3'), ... | | 著者 | Li, F.D, Xu, Z.Y. | | 登録日 | 2022-08-12 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | ZNF524 directly interacts with telomeric DNA and supports telomere integrity.
Nat Commun, 14, 2023
|
|
6XQT
 
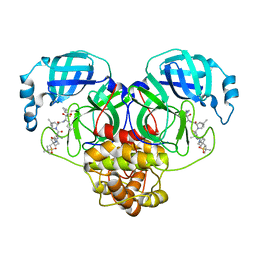 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Narlaprevir | | 分子名称: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kneller, D.W, Kovalevsky, A, Coates, L. | | 登録日 | 2020-07-10 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
7T1L
 
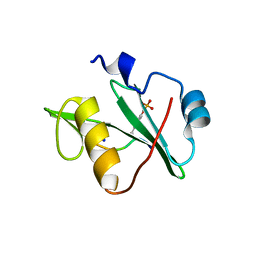 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | 分子名称: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | 著者 | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | 登録日 | 2021-12-02 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
