8E9R
 
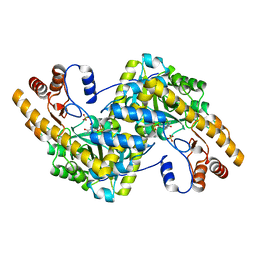 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
7TNH
 
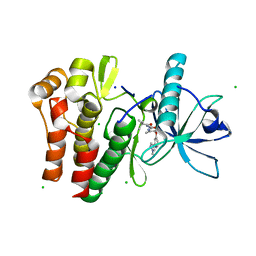 | | Crystal structure of CSF1R kinase domain in complex with DP-6233 | | 分子名称: | 2,2-dimethyl-N-[(6-methyl-5-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}pyridin-2-yl)carbamoyl]propanamide, CHLORIDE ION, Macrophage colony-stimulating factor 1 receptor,Fibroblast growth factor receptor 1 chimera, ... | | 著者 | Edwards, T.E, Arakaki, T.L, Chun, L, Flynn, D.L. | | 登録日 | 2022-01-21 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of acyl ureas as highly selective small molecule CSF1R kinase inhibitors.
Bioorg.Med.Chem.Lett., 74, 2022
|
|
4YDD
 
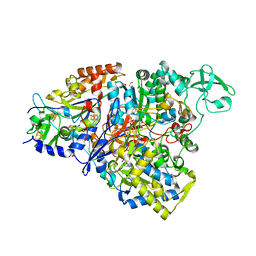 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | 登録日 | 2015-02-21 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
8E9L
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
4YFK
 
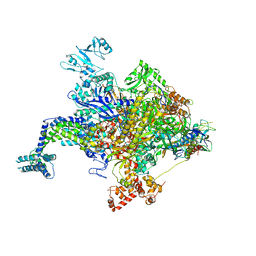 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | 分子名称: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | 登録日 | 2015-02-25 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.571 Å) | | 主引用文献 | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
7TUH
 
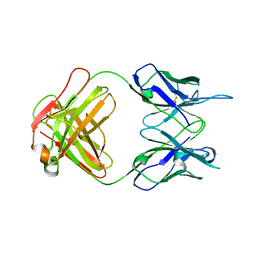 | | Crystal structure of anti-tapasin PaSta2-Fab | | 分子名称: | PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain | | 著者 | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | 登録日 | 2022-02-02 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | 著者 | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-08-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
8E9O
 
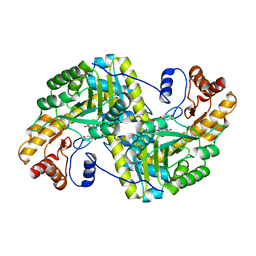 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
6QGP
 
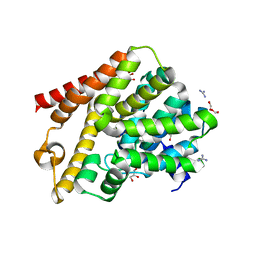 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769 | | 分子名称: | 1-cycloheptyl-3-[3-(cyclopentyloxy)-4-methoxyphenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | 登録日 | 2019-01-12 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769
To be published
|
|
6CW9
 
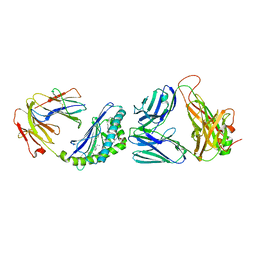 | | Structure of alpha-GC[8,16P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, J, Zajonc, D. | | 登録日 | 2018-03-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7OQP
 
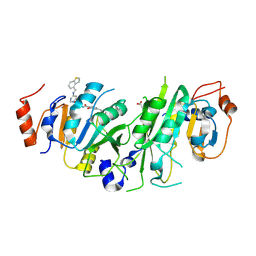 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ113 | | 分子名称: | ACETATE ION, N-[[(3R)-1-[6-(1-benzothiophen-4-ylmethylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-4-[[(3S)-3-propan-2-yl-2-azaspiro[3.3]heptan-2-yl]methyl]benzamide, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Huang, D, Caflisch, A. | | 登録日 | 2021-06-03 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
8E9T
 
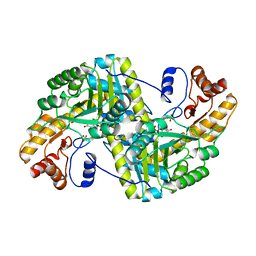 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9K
 
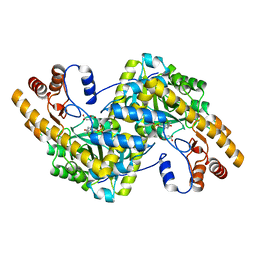 | | Crystal structure of wild-type E. coli aspartate aminotransferase bound to maleate at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9S
 
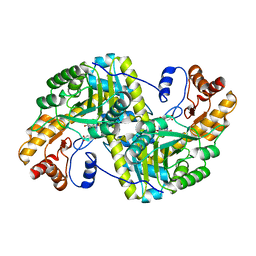 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
4YJ3
 
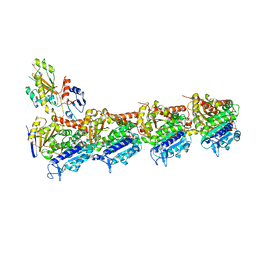 | | Crystal structure of tubulin bound to compound 2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(4-ethoxyphenyl)-3-(2-methoxyphenyl)-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazine, CALCIUM ION, ... | | 著者 | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | 登録日 | 2015-03-03 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.75 Å) | | 主引用文献 | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
8E9Q
 
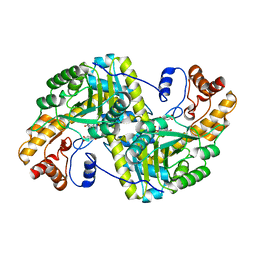 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
5JV5
 
 | |
5JVG
 
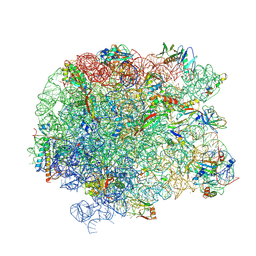 | | The large ribosomal subunit from Deinococcus radiodurans in complex with avilamycin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | 著者 | Krupkin, M, Wekselman, I, Matzov, D, Eyal, Z, Diskin Posner, Y, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | 登録日 | 2016-05-11 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.428 Å) | | 主引用文献 | Avilamycin and evernimicin induce structural changes in rProteins uL16 and CTC that enhance the inhibition of A-site tRNA binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6QBQ
 
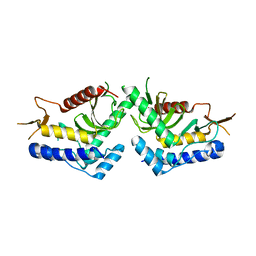 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200A S203A | | 分子名称: | Cell wall assembly regulator SMI1 | | 著者 | Guillien, M, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | 登録日 | 2018-12-21 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200A S203A
To Be Published
|
|
8IKS
 
 | |
6YQO
 
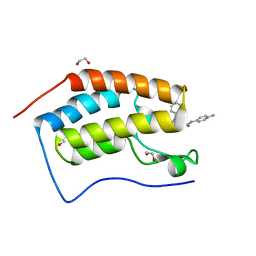 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | 分子名称: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | 著者 | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-04-17 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
8E9V
 
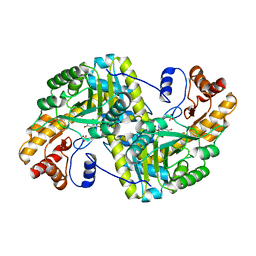 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
6YAC
 
 | | Plant PSI-ferredoxin supercomplex | | 分子名称: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Caspy, I, Nelson, N, Shkolnisky, Y, Klaiman, D, Sheinker, A. | | 登録日 | 2020-03-12 | | 公開日 | 2020-09-30 | | 最終更新日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | The structure of a triple complex of plant photosystem I with ferredoxin and plastocyanin.
Nat.Plants, 6, 2020
|
|
7OQL
 
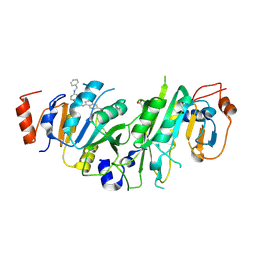 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ094 | | 分子名称: | (3~{R})-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]-3-[[[6-[[(3~{S})-3-propan-2-yl-2-azoniaspiro[3.3]heptan-2-yl]methyl]naphthalen-1-yl]amino]methyl]piperidin-3-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Huang, D, Caflisch, A. | | 登録日 | 2021-06-03 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | 分子名称: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | 著者 | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | 登録日 | 2017-03-09 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
