7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7MX1
 
 | | PLK-1 polo-box domain in complex with a high affinity macrocycle synthesized using a novel glutamic acid analog | | Descriptor: | ACE-PRO-LEU-ALA-SER-TPO, N-[(4S)-4,5-diamino-5-oxopentyl]-10-phenyldecanamide, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of a new orthogonally protected glutamic acid analog and its use in the preparation of high affinity polo-like kinase 1 polo-box domain - binding peptide macrocycles.
Org.Biomol.Chem., 19, 2021
|
|
6N93
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-nitronate-propionyl-oxa(dethia)-CoA | | Descriptor: | (2E)-2-(hydroxyimino)propanoic acid, IMIDAZOLE, Methylmalonyl-CoA decarboxylase, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
7N5B
 
 | | Structure of AtAtm3 in the outward-facing conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
6W7J
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ635p | | Descriptor: | 1,2-ETHANEDIOL, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
6XWP
 
 | |
5IRE
 
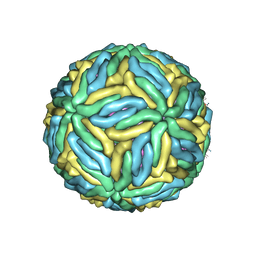 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
5I33
 
 | | Unligated adenylosuccinate synthetase from Cryptococcus neoformans | | Descriptor: | Adenylosuccinate synthetase | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
9BHK
 
 | | MerTK in complex with small molecule inhibitor 6-{1-[6-(3-hydroxy-3-methylbutoxy)-1,3-benzoxazol-2-yl]azetidin-3-yl}-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide | | Descriptor: | 6-{1-[6-(3-hydroxy-3-methylbutoxy)-1,3-benzoxazol-2-yl]azetidin-3-yl}-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | Jakob, C.G, Gurbani, D, Qiu, W. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Discovery of Potent Azetidine-Benzoxazole MerTK Inhibitors with In Vivo Target Engagement.
J.Med.Chem., 2024
|
|
4RP7
 
 | |
7N5A
 
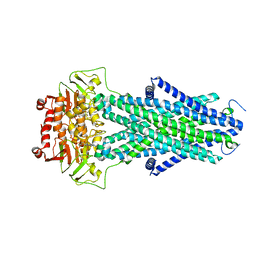 | | Structure of AtAtm3 in the closed conformation | | Descriptor: | ABC transporter B family member 25, mitochondrial, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Glutathione binding to the plant At Atm3 transporter and implications for the conformational coupling of ABC transporters.
Elife, 11, 2022
|
|
6TP6
 
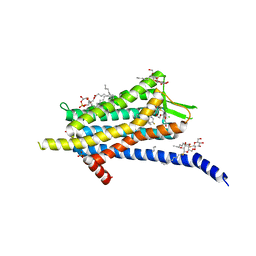 | | Crystal structure of the Orexin-1 receptor in complex with filorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CHLORIDE ION, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7N4T
 
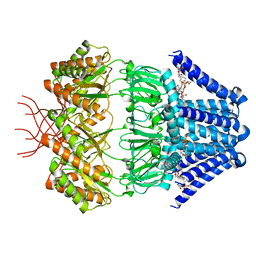 | | Low conductance mechanosensitive channel YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI | | Authors: | Catalano, C, Ben-Hail, D, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of Mechanosensitive Channel YnaI Using SMA2000: Challenges and Opportunities.
Membranes (Basel), 11, 2021
|
|
6TQ4
 
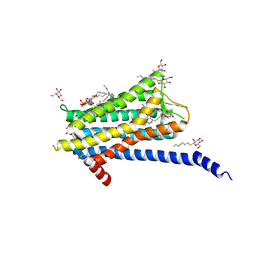 | | Crystal structure of the Orexin-1 receptor in complex with Compound 16 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-[1-(phenylsulfonyl)-1,8-diazaspiro[4.5]decan-8-yl]-1,3-benzoxazole, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5NKT
 
 | | FimA wt from E. coli | | Descriptor: | SULFATE ION, Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2017-04-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
5NKU
 
 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
3ITB
 
 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | Descriptor: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
4G21
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
8IO9
 
 | |
6YSI
 
 | | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
7TUG
 
 | | Crystal structure of Tapasin in complex with PaSta2-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
5AKM
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI G20S IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MG | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
6Z9C
 
 | | Structure of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability | | Descriptor: | Polymerase delta-interacting protein 2, SODIUM ION | | Authors: | Kulik, A.A, Maruszczak, K, Nabi, N.L.M, Bingham, R.J, Cooper, C.D.O. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability.
Protein Sci., 30, 2021
|
|
8OWD
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
5N9M
 
 | | Crystal structure of GatD - a glutamine amidotransferase from Staphylococcus aureus involved in peptidoglycan amidation | | Descriptor: | Cobyric acid synthase, GLUTAMINE, TETRAETHYLENE GLYCOL | | Authors: | Leisico, F, Vieira, D, Romao, M.R, Trincao, J, Santos-Silva, T. | | Deposit date: | 2017-02-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First insights of peptidoglycan amidation in Gram-positive bacteria - the high-resolution crystal structure of Staphylococcus aureus glutamine amidotransferase GatD.
Sci Rep, 8, 2018
|
|
