8AIT
 
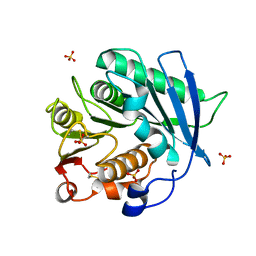 | | Crystal structure of cutinase PbauzCut from Pseudomonas bauzanensis | | Descriptor: | Cutinase, SULFATE ION | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
6H9G
 
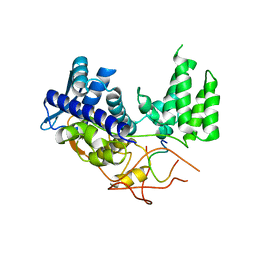 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1. | | Descriptor: | Nucleoprotein, Polypeptide loop | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Munier, S, Carlero, D, Naffakh, N, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-08-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6XUZ
 
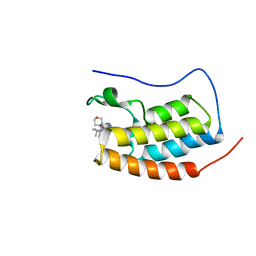 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3HSM
 
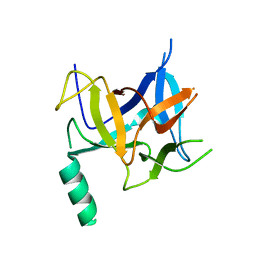 | | Crystal structure of distal N-terminal beta-trefoil domain of Ryanodine Receptor type 1 | | Descriptor: | Ryanodine receptor 1 | | Authors: | Amador, F.J, Liu, S, Ishiyama, N, Plevin, M.J, Wilson, A, MacLennan, D.H, Ikura, M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation "hot spot" loop
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6XWQ
 
 | |
8F7N
 
 | | Crystal structure of chemoreceptor McpZ ligand sensing domain | | Descriptor: | Methyl-accepting chemotaxis protein, YTTERBIUM (III) ION | | Authors: | Salar, S, Schubot, F.D. | | Deposit date: | 2022-11-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural analysis of the periplasmic domain of Sinorhizobium meliloti chemoreceptor McpZ reveals a novel fold and suggests a complex mechanism of transmembrane signaling.
Proteins, 91, 2023
|
|
6V36
 
 | |
6N5H
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 5 | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriano, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors
Int. J. Biol. Macromol., 129, 2019
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7SEE
 
 | | Structure of E. coli LetB delta (Ring6) mutant, Ring1 in the closed state (Model 1) | | Descriptor: | MCE family protein, Intermembrane transport protein YebT chimera, UNKNOWN ATOM OR ION | | Authors: | Vieni, C, Coudray, N, Bhabha, G, Ekiert, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Role of Ring6 in the Function of the E. coli MCE Protein LetB.
J.Mol.Biol., 434, 2022
|
|
6N6H
 
 | | Vibrio cholerae Oligoribonuclease bound to pCpU | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*CP*U)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6PL4
 
 | | TRK-A IN COMPLEX WITH LIGAND 1 | | Descriptor: | High affinity nerve growth factor receptor, N-{[5-(methoxymethyl)-2-(trifluoromethoxy)phenyl]methyl}-N'-(8-methyl-2-phenylimidazo[1,2-a]pyrazin-3-yl)urea | | Authors: | Subramanian, G, Brown, D.G. | | Deposit date: | 2019-06-30 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | In Pursuit of an Allosteric Human Tropomyosin Kinase A (hTrkA) Inhibitor for Chronic Pain
Acs Med.Chem.Lett., 2021
|
|
8FD9
 
 | |
5KEM
 
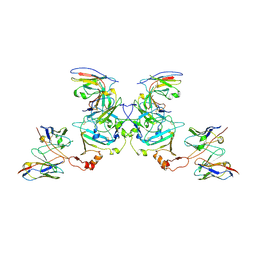 | | EBOV sGP in complex with variable Fab domains of IgGs c13C6 and BDBV91 | | Descriptor: | BDBV91 variable Fab domain heavy chain, BDBV91 variable Fab domain light chain, Ebola secreted glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
6WDY
 
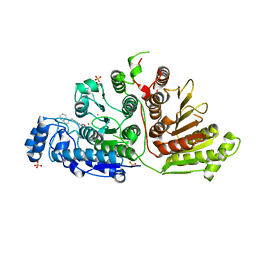 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with Indole Phenylhydroxamate Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-hydroxy-4-[(1H-indol-1-yl)methyl]benzamide, PHOSPHATE ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for the Selective Inhibition of HDAC10, the Cytosolic Polyamine Deacetylase.
Acs Chem.Biol., 15, 2020
|
|
6WDW
 
 | |
7QPW
 
 | |
6WEP
 
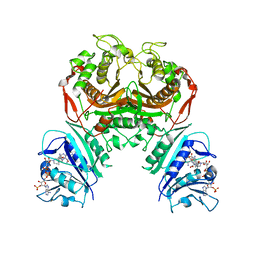 | | Crystal structure of TS-DHFR from Cryptosporidium hominis with Apo-TS site | | Descriptor: | Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Czyzyk, D.J, Ruiz, V.G, Kumar, V.P, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Targeting the TS dimer interface in bifunctional Cryptosporidium hominis TS-DHFR from parasitic protozoa: Virtual screening identifies novel TS allosteric inhibitor
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4RE0
 
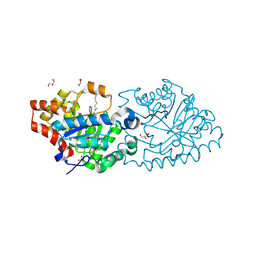 | | Crystal structure of VmoLac in P622 space group | | Descriptor: | COBALT (II) ION, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | Deposit date: | 2014-09-20 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
6WXW
 
 | |
6PAR
 
 | |
3ML3
 
 | | Crystal structure of the IcsA autochaperone region | | Descriptor: | Outer membrane protein icsA autotransporter | | Authors: | Diezmann, D, Kuhnel, K. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Autochaperone Region from the Shigella flexneri Autotransporter IcsA
J.Bacteriol., 193, 2011
|
|
7QPY
 
 | |
6IO0
 
 | | Human IDH1 R132C mutant complexed with compound A. | | Descriptor: | (2E)-3-{3-[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazole-4-carbonyl]-1-methyl-1H-indol-7-yl}prop-2-enoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Suzuki, M, Baba, D, Hanzawa, H. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Potent Blood-Brain Barrier-Permeable Mutant IDH1 Inhibitor Suppresses the Growth of Glioblastoma with IDH1 Mutation in a Patient-Derived Orthotopic Xenograft Model.
Mol.Cancer Ther., 19, 2020
|
|
6YMO
 
 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pS617 peptide | | Descriptor: | 14-3-3 protein zeta/delta, 2-HYDROXYBENZOIC ACID, Glucocorticoid receptor, ... | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
