5J8G
 
 | | Structure of nitroreductase from E. cloacae complexed with para-nitrobenzoic acid | | Descriptor: | 4-NITROBENZOIC ACID, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.-F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
6PRK
 
 | | X-ray Crystal Structure of Bacillus subtilis RicA in complex with RicF | | Descriptor: | RicA, RicF | | Authors: | Khaja, F.T, Jeffrey, P.D, Neiditch, M.B, Dubnau, D. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Function Studies of the Bacillus subtilis Ric Proteins Identify the Fe-S Cluster-Ligating Residues and Their Roles in Development and RNA Processing.
Mbio, 10, 2019
|
|
6GDH
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3') | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
7P1K
 
 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6VRG
 
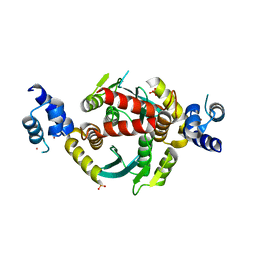 | | Structure of HIV-1 integrase with native amino-terminal sequence | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Eilers, G, Gupta, K, Allen, A, Zhou, J, Hwang, Y, Cory, M, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2020-02-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of the amino-terminal sequence on the structure and function of HIV integrase.
Retrovirology, 17, 2020
|
|
7BLN
 
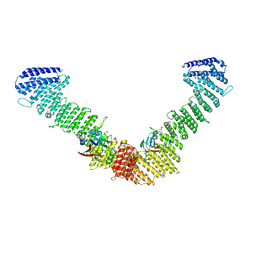 | | VPS35/VPS29 arch of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
6RCO
 
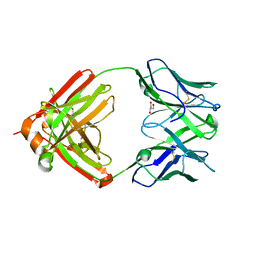 | | PfRH5-binding monoclonal antibody R5.004 | | Descriptor: | ACETATE ION, GLYCEROL, R5.004 heavy chain, ... | | Authors: | Alanine, D.G.W, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6NET
 
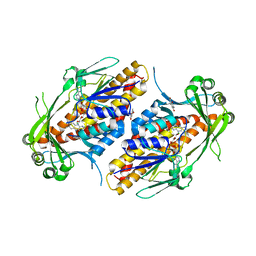 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
8CYK
 
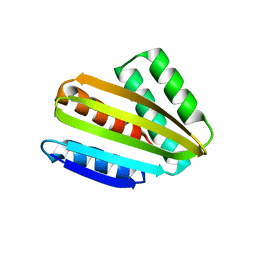 | | Crystal structure of hallucinated protein HALC1_878 | | Descriptor: | HALC1_878 | | Authors: | Ragotte, R.J, Bera, A.K, Milles, L.F, Wicky, B.I.M, Baker, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Robust deep learning-based protein sequence design using ProteinMPNN.
Science, 378, 2022
|
|
4WZ5
 
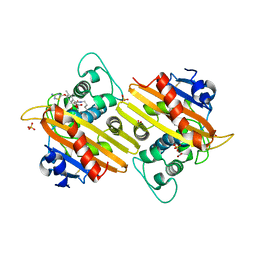 | | Crystal structure of P. aeruginosa OXA10 | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
Acs Infect Dis., 1, 2015
|
|
8DDG
 
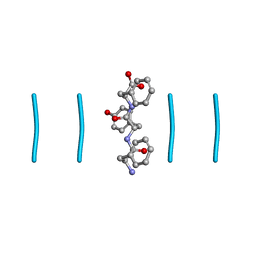 | |
8D9C
 
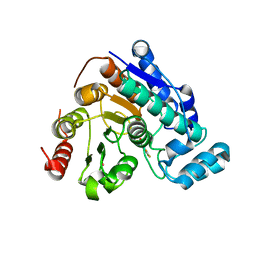 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 10 | | Descriptor: | 2,3,4,5,6-pentafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D08
 
 | | Hallucinated C4 protein assembly HALC4_135 | | Descriptor: | HALC4_135 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
7TLF
 
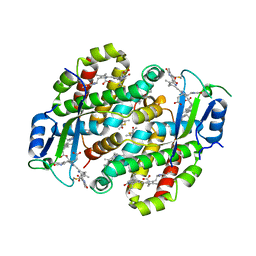 | | Structure of the photoacclimated Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOERYTHROBILIN, Phycoerythrin alpha-subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
8D99
 
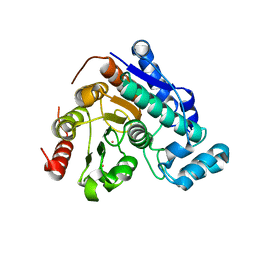 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 7 | | Descriptor: | 2,3,6-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9B
 
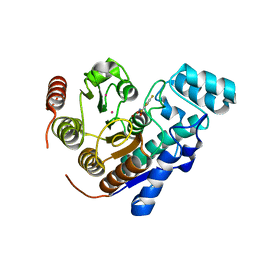 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 9 | | Descriptor: | 2,3,5,6-tetrafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8OUO
 
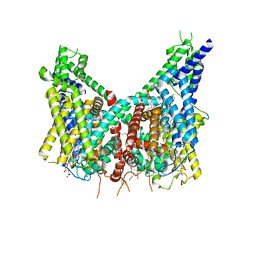 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 2024
|
|
8D09
 
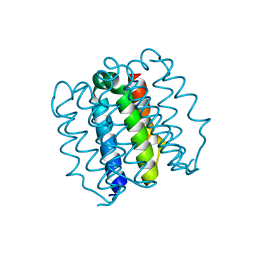 | | Hallucinated C4 protein assembly HALC4_136 | | Descriptor: | HALC4_136 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D98
 
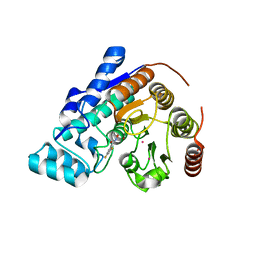 | |
8CTB
 
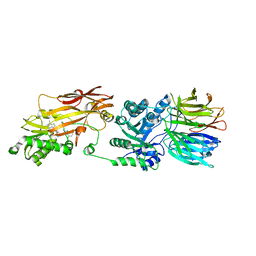 | | Human PRMT5:MEP50 structure with Fragment 3 and MTA Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 7-chloro-1-methyl-1H-benzimidazol-2-amine, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
8D3B
 
 | |
6NDE
 
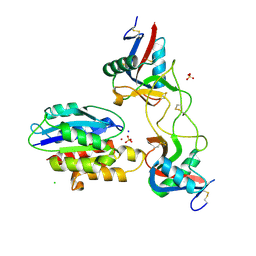 | |
8D9A
 
 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 8 | | Descriptor: | 2,3,5-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Christianson, D.W. | | Deposit date: | 2022-06-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
7ZVT
 
 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
