5D9M
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG | | Descriptor: | B-1,4-endoglucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
5D9P
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG | | Descriptor: | B-1,4-endoglucanase, CALCIUM ION, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-N-acetyl-beta-D-glucopyranosylamine | | Authors: | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
6UVZ
 
 | | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis | | Descriptor: | Amidohydrolase 2, CITRIC ACID, NONAETHYLENE GLYCOL | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-11-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis
To Be Published
|
|
4NO1
 
 | | yCP in complex with Z-Leu-Leu-Leu-B(OH)2 | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NO6
 
 | | yCP in complex with Z-Leu-Leu-Leu-vinylsulfone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(3S)-5-methyl-1-(methylsulfonyl)hexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NNN
 
 | | yCP in complex with MG132 | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-18 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MWA
 
 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4O2I
 
 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
4NO8
 
 | | yCP in complex with Z-Leu-Leu-Leu-ketoamide | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S,3S)-2-hydroxy-5-methyl-1-oxo-1-(phenylamino)hexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NO9
 
 | | yCP in complex with Z-Leu-Leu-Leu-epoxyketone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-L-leucinamide, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(1R,2S)-1-hydroxy-4-methyl-1-[(2R)-2-methyloxiran-2-yl]pentan-2-yl}-L-leucinamide, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NNW
 
 | | yCP in complex with Z-Leu-Leu-Leu-ketoaldehyde | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2R,3S)-1,2-dihydroxy-5-methylhexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O8O
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to alpha-L-arabinose | | Descriptor: | Alpha-L-arabinofuranosidase, CALCIUM ION, alpha-L-arabinofuranose | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
3HE1
 
 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
3HIU
 
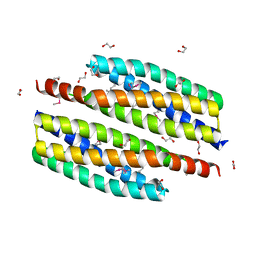 | | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
3ICF
 
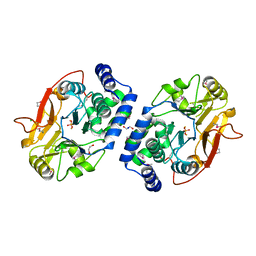 | | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Cui, H, Kagan, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5
To be Published
|
|
3IBZ
 
 | | Crystal structure of putative tellurium resistant like protein (TerD) from Streptomyces coelicolor A3(2) | | Descriptor: | CALCIUM ION, Putative tellurium resistant like protein TerD, SULFATE ION | | Authors: | Klimecka, M, Chruszcz, M, Cymborowski, M, Xu, X, Cui, H, Joachimiak, A, Edwards, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-18 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of putative tellurium resistant like protein (TerD) from Streptomyces coelicolor A3(2)
To be Published
|
|
3IV4
 
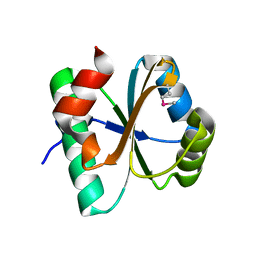 | | A putative oxidoreductase with a thioredoxin fold | | Descriptor: | Putative oxidoreductase | | Authors: | Fan, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a putative oxidoreductase with a thioredoxin fold
To be Published
|
|
3I7M
 
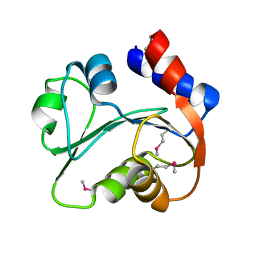 | | N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis. | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis.
To be Published
|
|
3IC4
 
 | | The crystal structure of the glutaredoxin(grx-1) from Archaeoglobus fulgidus | | Descriptor: | Glutaredoxin (Grx-1), MAGNESIUM ION | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the glutaredoxin from Archaeoglobus fulgidus
To be Published
|
|
3JU1
 
 | | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein | | Descriptor: | ACETIC ACID, Enoyl-CoA hydratase/isomerase family protein, FORMIC ACID, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein
To be Published
|
|
3K0B
 
 | | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, predicted N6-adenine-specific DNA methylase | | Authors: | Nocek, B, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365
To be Published
|
|
3K6H
 
 | | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase family protein, SULFATE ION | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58
To be Published
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4EW5
 
 | | C-terminal domain of inner membrane protein CigR from Salmonella enterica. | | Descriptor: | 1,2-ETHANEDIOL, CigR Protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Niemann, G.S, Merkley, E.D, Savchenko, A, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | C-terminal domain of inner membrane protein CigR from Salmonella enterica.
To be Published
|
|
3KWP
 
 | | Crystal structure of putative methyltransferase from Lactobacillus brevis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Predicted methyltransferase | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-01 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of putative methyltransferase from Lactobacillus brevis
To be Published
|
|
