2H2I
 
 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
2H2G
 
 | |
2H2D
 
 | |
2H2F
 
 | |
2H2H
 
 | |
1E7Y
 
 | | ACTIVE SITE MUTANT (D177->N) OF GLUCOSE 6-PHOSPHATE DEHYDROGENASE FROM LEUCONOSTOC MESENTEROIDES COMPLEXED WITH SUBSTRATE AND NADPH | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CALCIUM ION, GLUCOSE 6-PHOSPHATE 1-DEHYDROGENASE, ... | | Authors: | Adams, M.J, Cosgrove, M.S, Gover, S. | | Deposit date: | 2000-09-12 | | Release date: | 2000-12-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | An Examination of the Role of Asp-177 in the His-Asp Catalytic Dyad of Leuconostoc Mesenteroides Glucose 6-Phosphate Dehydrogenase: X-Ray Structure and Ph Dependence of Kinetic Parameters of the D177N Mutant Enzyme
Biochemistry, 39, 2000
|
|
1E7M
 
 | |
7KKF
 
 | |
5SXM
 
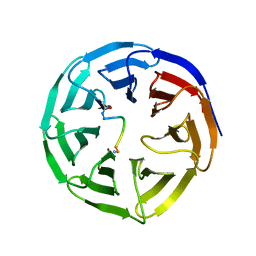 | | WDR5 in complex with MLL Win motif peptidomimetic | | Descriptor: | ACE-ALA-ARG-THR-GLU-VAL-TYR-NH2, WD repeat-containing protein 5 | | Authors: | Alicea-Velazquez, N.L, Shinsky, S.A, Cosgrove, M.S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeted Disruption of the Interaction between WD-40 Repeat Protein 5 (WDR5) and Mixed Lineage Leukemia (MLL)/SET1 Family Proteins Specifically Inhibits MLL1 and SETd1A Methyltransferase Complexes.
J.Biol.Chem., 291, 2016
|
|
1MA3
 
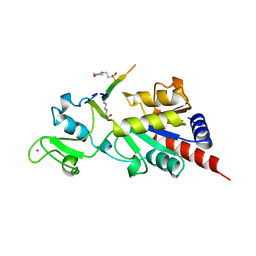 | | Structure of a Sir2 enzyme bound to an acetylated p53 peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular tumor antigen p53, Transcriptional regulatory protein, ... | | Authors: | Avalos, J.L, Celic, I, Muhammad, S, Cosgrove, M.S, Boeke, J.D, Wolberger, C. | | Deposit date: | 2002-07-31 | | Release date: | 2002-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 enzyme bound to an acetylated p53 peptide
Mol.Cell, 10, 2002
|
|
3EG6
 
 | |
4ES0
 
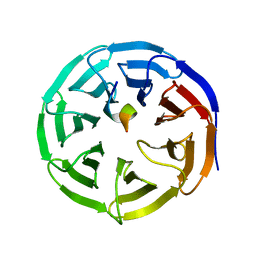 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERZ
 
 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERQ
 
 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ESG
 
 | | X-ray structure of WDR5-MLL1 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4EWR
 
 | | X-ray structure of WDR5-SETd1a Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-27 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
