6IWB
 
 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | Descriptor: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | Authors: | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
9FKD
 
 | |
8S1X
 
 | |
7AYE
 
 | | Crystal structure of the computationally designed chemically disruptable heterodimer LD6-MDM2 | | Descriptor: | Isoform 11 of E3 ubiquitin-protein ligase Mdm2, Thiol:disulfide interchange protein DsbD | | Authors: | Yang, C, Lau, K, Pojer, F, Correia, B.E. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A rational blueprint for the design of chemically-controlled protein switches.
Nat Commun, 12, 2021
|
|
6XXV
 
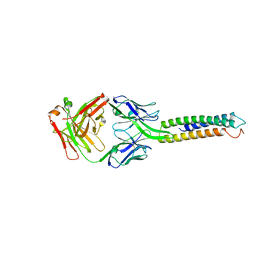 | | Crystal Structure of a computationally designed Immunogen S2_1.2 in complex with its elicited antibody C57 | | Descriptor: | Antibody C57, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Correia, B.E, Pojer, F. | | Deposit date: | 2020-01-28 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20116425 Å) | | Cite: | De novo protein design enables the precise induction of RSV-neutralizing antibodies.
Science, 368, 2020
|
|
6YWC
 
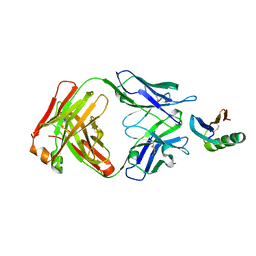 | | De novo designed protein 4E1H_95 in complex with 101F antibody | | Descriptor: | Antibody 101F, Heavy Chain, light chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
6YWD
 
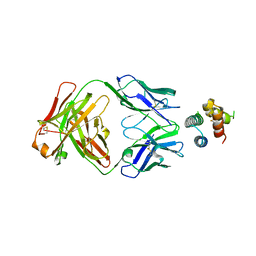 | | De novo designed protein 4H_01 in complex with Mota antibody | | Descriptor: | Antibody Mota, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
8P49
 
 | |
8QHP
 
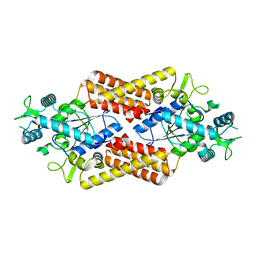 | |
8Q70
 
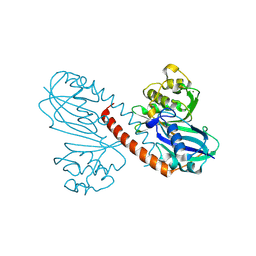 | |
8OYS
 
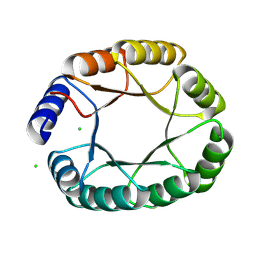 | | De novo designed TIM barrel fold TBF_24 | | Descriptor: | CHLORIDE ION, De novo designed TIM-barrel | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYV
 
 | | De novo designed Claudin fold CLF_4 | | Descriptor: | De novo designed soluble Claudin | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYY
 
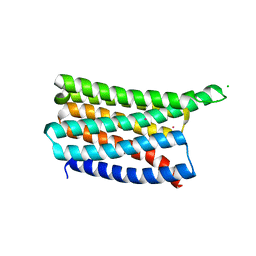 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYW
 
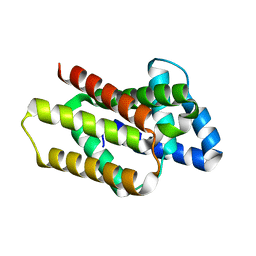 | |
