4GVZ
 
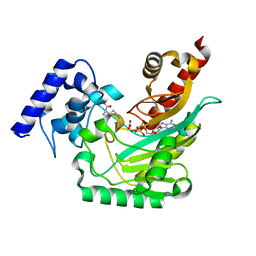 | | Crystal structure of arginine kinase in complex with D-arginine, MgADP, and nitrate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
5E0L
 
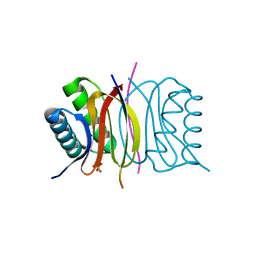 | | LC8 - Chica (415-424) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
5E0M
 
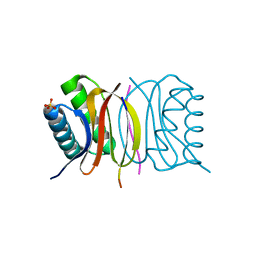 | | LC8 - Chica (468-476) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
8TKP
 
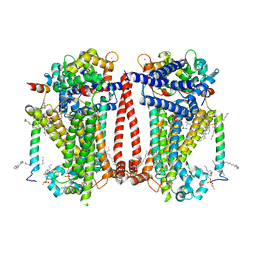 | | Structure of the C. elegans TMC-2 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4GW0
 
 | | Crystal structure of arginine kinase in complex with imino-L-ornithine, MgADP, and nitrate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, MAGNESIUM ION, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
4GW2
 
 | | Crystal structure of arginine kinase in complex with L-ornithine, MgADP, and nitrate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, L-ornithine, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
4GVY
 
 | | Crystal structure of arginine kinase in complex with L-citrulline and MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, CITRULLINE, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
6J7W
 
 | | Crystal Structure of Human BCMA in complex with UniAb(TM) VH | | Descriptor: | Tumor necrosis factor receptor superfamily member 17, UniAb | | Authors: | Clarke, S.C, Ma, B, Trinklein, N.D, Schellenberger, U, Osborn, M, Ouisse, L, Boudreau, A, Davison, L, Harris, K.E, Ugamraj, H, Balasubramani, A, Dang, K, Jorgensen, B, Ogana, H, Pham, D, Pratap, P, Sankaran, P, Anegon, I, van Schooten, W, Bruggemann, M, Buelow, R, Force Aldred, S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multispecific Antibody Development Platform Based on Human Heavy Chain Antibodies
Front Immunol, 9, 2018
|
|
7SN0
 
 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Clark, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
 | |
7KFV
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B12 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody C1A-B12 Fab, Spike glycoprotein, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFW
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B3 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of antibody C1A-B3 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFY
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-F10 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-F10 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFX
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-C2 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-C2 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
4LMQ
 
 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
4URJ
 
 | | Crystal structure of human BJ-TSA-9 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN FAM83A | | Authors: | Pinkas, D.M, Sanvitale, C, Wang, D, Krojer, T, Kopec, J, Chaikuad, A, Dixon Clarke, S, Berridge, G, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of Human Bj-Tsa-9
To be Published
|
|
7USY
 
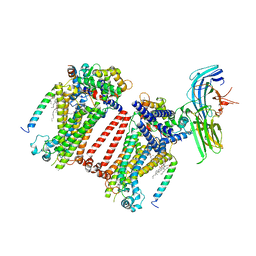 | | Structure of C. elegans TMC-1 complex with ARRD-6 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USX
 
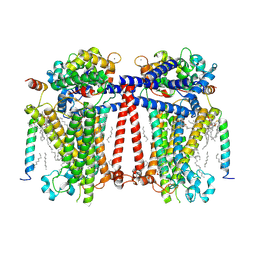 | | Structure of Contracted C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USW
 
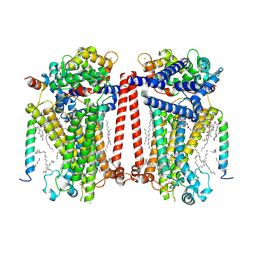 | | Structure of Expanded C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
1P50
 
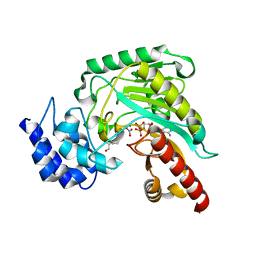 | | Transition state structure of an Arginine Kinase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1P52
 
 | | Structure of Arginine kinase E314D mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
7ABL
 
 | | HBV pgRNA T=4 NCP icosahedral symmetry | | Descriptor: | Capsid protein | | Authors: | Patel, N, Clark, S, Weis, E.U, Mata, C.P, Bohon, J, Farquhar, E, Ranson, N.A, Twarock, R, Stockley, P.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro functional analysis of gRNA sites regulating assembly of hepatitis B virus.
Commun Biol, 4, 2021
|
|
2MVF
 
 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
3JPZ
 
 | | Crystal Structure of Lombricine Kinase | | Descriptor: | Lombricine kinase, NITRATE ION | | Authors: | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
3JQ3
 
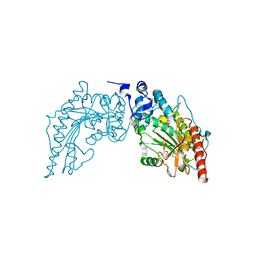 | | Crystal Structure of Lombricine Kinase, complexed with substrate ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lombricine kinase | | Authors: | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2009-09-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
