5HAW
 
 | |
2XJ2
 
 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ0
 
 | | Protein kinase Pim-1 in complex with fragment-4 from crystallographic fragment screen | | Descriptor: | (E)-3-(2-AMINO-PYRIDINE-5YL)-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ1
 
 | | Protein kinase Pim-1 in complex with small molecule inibitor | | Descriptor: | (2E)-3-(3-{6-[(TRANS-4-AMINOCYCLOHEXYL)AMINO]PYRAZIN-2-YL}PHENYL)PROP-2-ENOIC ACID, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIZ
 
 | | Protein kinase Pim-1 in complex with fragment-3 from crystallographic fragment screen | | Descriptor: | (E)-PYRIDIN-4-YL-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIX
 
 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1QX5
 
 | |
1SDL
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
2LIE
 
 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
4MCT
 
 | |
2MK1
 
 | |
1BIW
 
 | | DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS | | Descriptor: | CALCIUM ION, N1-HYDROXY-2-(3-HYDROXY-PROPYL)-3-ISOBUTYL-N4-[1-(2-METHOXY-ETHYL)-2-OXO-AZEPAN-3-YL]-SUCCINAMIDE, PROTEIN (STROMELYSIN-1 COMPLEX), ... | | Authors: | Natchus, M.G, Cheng, M, Wahl, C.T, Pikul, S, Almstead, N.G, Bradley, R.S, Taiwo, Y.O, Mieling, G.E, Dunaway, C.M, Snider, C.E, McIver, J.M, Barnett, B.L, McPhail, S.J, Anastasio, M.B, De, B. | | Deposit date: | 1998-06-19 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of conformationally-constrained MMP inhibitors.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
4R4E
 
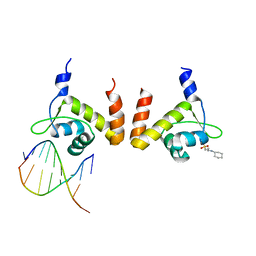 | | Structure of GlnR-DNA complex | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DNA (5'-D(*AP*TP*TP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*TP*A)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
3MKY
 
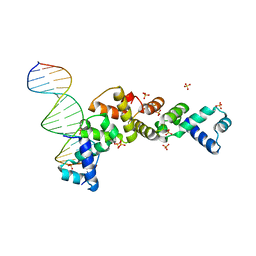 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
1JT0
 
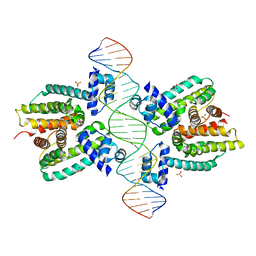 | | Crystal structure of a cooperative QacR-DNA complex | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, QACA operator, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-20 | | Release date: | 2002-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR.
EMBO J., 21, 2002
|
|
1JUP
 
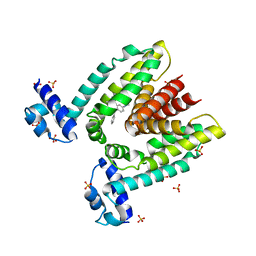 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to malachite green | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, MALACHITE GREEN, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1SXI
 
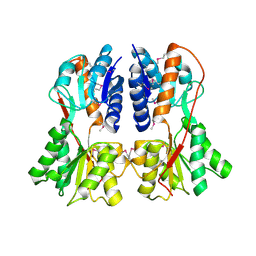 | | Structure of apo transcription regulator B. megaterium | | Descriptor: | Glucose-resistance amylase regulator, MAGNESIUM ION | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1JTX
 
 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to crystal violet | | Descriptor: | CRYSTAL VIOLET, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
4I5B
 
 | |
4OB4
 
 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
3TPV
 
 | | Structure of pHipA bound to ADP | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T.M, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
1JTY
 
 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to ethidium | | Descriptor: | ETHIDIUM, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JUM
 
 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to the natural drug berberine | | Descriptor: | BERBERINE, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JUS
 
 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to rhodamine 6G | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, RHODAMINE 6G, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-27 | | Release date: | 2001-12-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
3PM1
 
 | | Structure of QacR E90Q bound to Ethidium | | Descriptor: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
