4FRA
 
 | | Crystal Structure of ABBA+UDP+Gal at pH 5.0 with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
4FRE
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 6.5 with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
4FRM
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 7.0 with MPD as the cryoprotectant | | Descriptor: | Histo-blood group ABO system transferase, URIDINE-5'-DIPHOSPHATE, beta-D-galactopyranose | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
8PJN
 
 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | Descriptor: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8QBN
 
 | |
4GSL
 
 | | Crystal structure of an Atg7-Atg3 crosslinked complex | | Descriptor: | Autophagy-related protein 3, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Kaiser, S.E, Mao, K, Taherbhoy, A.M, Yu, S, Olszewski, J.L, Duda, D.M, Kurinov, I, Deng, A, Fenn, T.D, Klionsky, D.J, Schulman, B.A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2AST
 
 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
4GAO
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | BROMIDE ION, DCN1-like protein 2, NEDD8-conjugating enzyme Ubc12 | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-25 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
4RG6
 
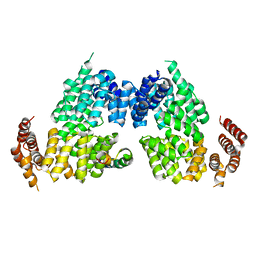 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
1ZFN
 
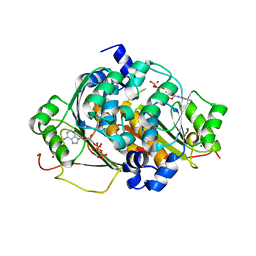 | | Structural Analysis of Escherichia coli ThiF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylyltransferase thiF, ZINC ION | | Authors: | Duda, D.M, Walden, H, Sfondouris, J, Schulman, B.A. | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Analysis of Escherichia Coli ThiF.
J.Mol.Biol., 349, 2005
|
|
4RG9
 
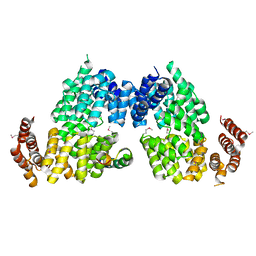 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
1ZKM
 
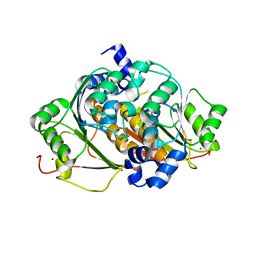 | | Structural Analysis of Escherichia Coli ThiF | | Descriptor: | Adenylyltransferase thiF, ZINC ION | | Authors: | Duda, D.M, Walden, H, Sfondouris, J, Schulman, B.A. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Analysis of Escherichia Coli ThiF.
J.Mol.Biol., 349, 2005
|
|
4GSK
 
 | | Crystal structure of an Atg7-Atg10 crosslinked complex | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, Ubiquitin-like-conjugating enzyme ATG10, ZINC ION | | Authors: | Kaiser, S.E, Mao, K, Taherbhoy, A.M, Yu, S, Olszewski, J.L, Duda, D.M, Kurinov, I, Deng, A, Fenn, T.D, Klionsky, D.J, Schulman, B.A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2PGY
 
 | | GTB C209A, no Hg | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase | | Authors: | Letts, J.A, Schuman, B. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The effect of heavy atoms on the conformation of the active-site polypeptide loop in human ABO(H) blood-group glycosyltransferase B.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2PGV
 
 | | GTB C209A | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION | | Authors: | Letts, J.A, Schuman, B. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The effect of heavy atoms on the conformation of the active-site polypeptide loop in human ABO(H) blood-group glycosyltransferase B.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1Y8X
 
 | | Structural basis for recruitment of Ubc12 by an E2-binding domain in NEDD8's E1 | | Descriptor: | Ubiquitin-activating enzyme E1C, Ubiquitin-conjugating enzyme E2 M | | Authors: | Huang, D.T, Paydar, A, Zhuang, M, Waddell, M.B, Holton, J.M, Schulman, B.A. | | Deposit date: | 2004-12-13 | | Release date: | 2005-02-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1.
Mol.Cell, 17, 2005
|
|
1TT5
 
 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4GBA
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | DCN1-like protein 3, NEDD8-conjugating enzyme UBE2F | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
5AN8
 
 | | Cryo-electron microscopy structure of rabbit TRPV2 ion channel | | Descriptor: | TRPV2 | | Authors: | Zubcevic, L, Herzik, M.A.J, Chung, B.C, Lander, G.C, Lee, S.Y. | | Deposit date: | 2015-09-04 | | Release date: | 2015-12-23 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-Electron Microscopy of the Trpv2 Ion Channel
Nat.Struct.Mol.Biol., 23, 2016
|
|
7KTX
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
3RTR
 
 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1YOV
 
 | |
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | Descriptor: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
7KWA
 
 | | Structure of DCN1 bound to N-((4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-(trifluoromethyl)benzamide | | Descriptor: | Endolysin,DCN1-like protein 1, N-[(4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-(trifluoromethyl)benzamide | | Authors: | Kim, H.S, Hammill, J.T, Schulman, B.A, Guy, R.K, Scott, D.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.
J.Med.Chem., 64, 2021
|
|
5BTV
 
 | | Crystal structure of human 14-3-3 sigma in complex with a Tau-protein peptide surrounding pS324 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ottmann, C, Schumacher, B, Bartel, M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-20 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau.
Faseb J., 29, 2015
|
|
