8EYX
 
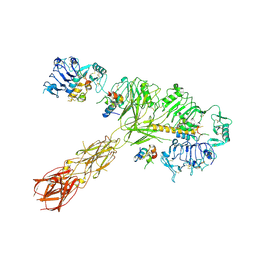 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Asymmetric conformation 1 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYY
 
 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S, denoted as IR-3CS) Asymmetric conformation 2 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6PXW
 
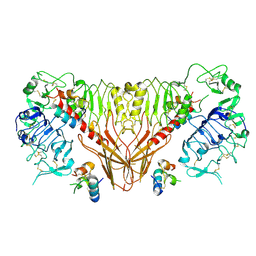 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
5KJ0
 
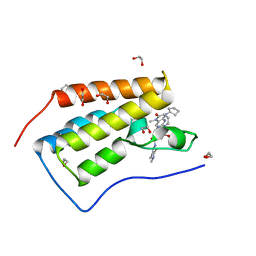 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH DB-1-264-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(7~{R})-8-cyclopentyl-7-ethyl-5-methyl-6-oxidanylidene-7~{H}-pteridin-2-yl]-methyl-amino]-3-methoxy-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W, Schonbrunn, E. | | Deposit date: | 2016-06-17 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assessment of Bromodomain Target Engagement by a Series of BI2536 Analogues with Miniaturized BET-BRET.
ChemMedChem, 11, 2016
|
|
7BJY
 
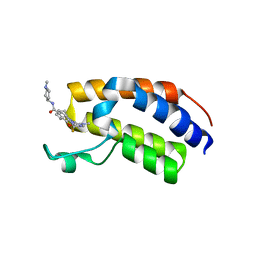 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to Ro3280 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Isoform 2 of Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8CZA
 
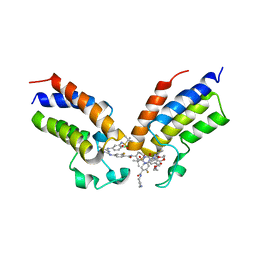 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to GXH-IV-075 | | Descriptor: | 4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluoro-N-[1-(14-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]phenyl}-14-oxo-4,7,10-trioxa-13-azatetradecanan-1-oyl)piperidin-4-yl]benzamide, Bromodomain testis-specific protein, SODIUM ION | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6B5H
 
 | |
6B5G
 
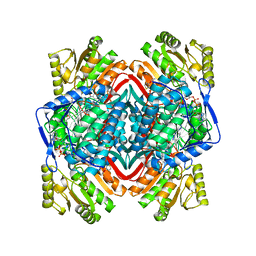 | |
6B5I
 
 | |
5KDH
 
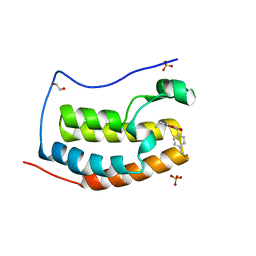 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH A DIHYDROPYRIDOPYRIMIDINE SCAFFOLD INHIBITOR | | Descriptor: | (5~{S})-1-ethyl-5-(4-methylphenyl)-8,9-dihydro-5~{H}-furo[3,4]pyrido[3,5-~{b}]pyrimidine-2,4,6-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2016-06-08 | | Release date: | 2017-08-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BET Bromodomain Inhibitors with One-Step Synthesis Discovered from Virtual Screen.
J. Med. Chem., 60, 2017
|
|
6PPA
 
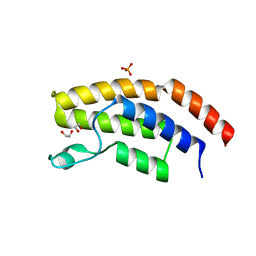 | | Crystal structure of the unliganded bromodomain of human BRD7 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, PHOSPHATE ION | | Authors: | Chan, A, Karim, M.R, Zhu, J, Schonbrunn, E. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
4ZO1
 
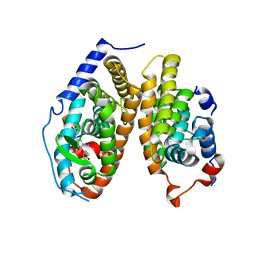 | | Crystal Structure of the T3-bound TR-beta Ligand-binding Domain in complex with RXR-alpha | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha, ... | | Authors: | Bruning, J.B, Kojetin, D.J, Matta-Camacho, E, Hughes, T.S, Srinivasan, S, Nwachukwu, J.C, Cavett, V, Nowak, J, Chalmers, M.J, Marciano, D.P, Kamenecka, T.M, Rance, M, Shulman, A.I, Mangelsdorf, D.J, Griffin, P.R, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Structural mechanism for signal transduction in RXR nuclear receptor heterodimers.
Nat Commun, 6, 2015
|
|
5NUS
 
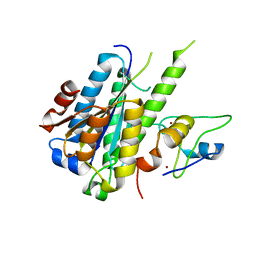 | | Structure of a minimal complex between p44 and p34 from Chaetomium thermophilum | | Descriptor: | ZINC ION, p34, p44 | | Authors: | Koelmel, W, Schoenwetter, E, Kuper, J, Schmitt, D.R, Kisker, C. | | Deposit date: | 2017-05-02 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
3NY1
 
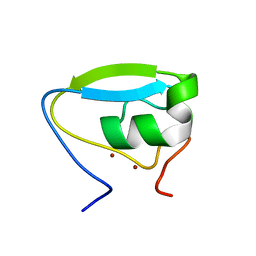 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY3
 
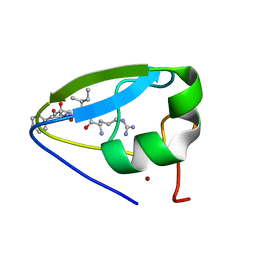 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8EYR
 
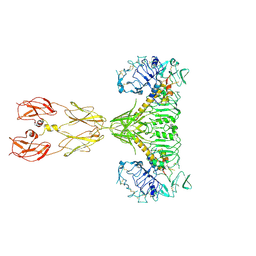 | | Cryo-EM structure of two IGF1 bound full-length mouse IGF1R mutant (four glycine residues inserted in the alpha-CT; IGF1R-P674G4): symmetric conformation | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
6BNT
 
 | |
6PYH
 
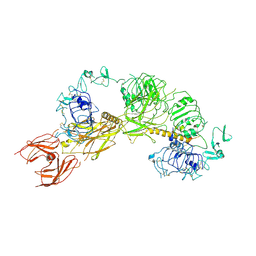 | | Cryo-EM structure of full-length IGF1R-IGF1 complex. Only the extracellular region of the complex is resolved. | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Choi, E, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of the activation of type 1 insulin-like growth factor receptor.
Nat Commun, 10, 2019
|
|
5VD1
 
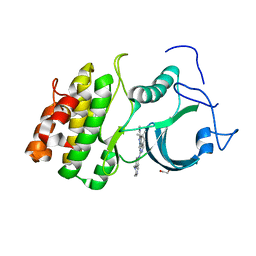 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH PHA-848125 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, N,1,4,4-TETRAMETHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VCW
 
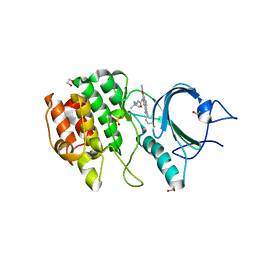 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VCZ
 
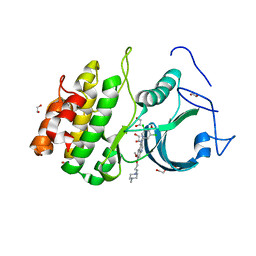 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Bosutinib isomer | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3,5-DICHLORO-4-METHOXYPHENYL)AMINO]-6-METHOXY-7-[3-(4-METHYLPIPERAZIN-1-YL)PROPOXY]QUINOLINE-3-CARBONITRILE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VCX
 
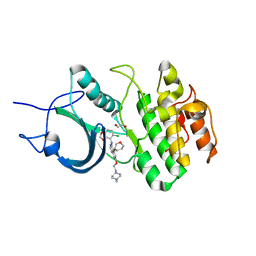 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN (UNTREATED) IN COMPLEX WITH SARACATINIB | | Descriptor: | 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VD3
 
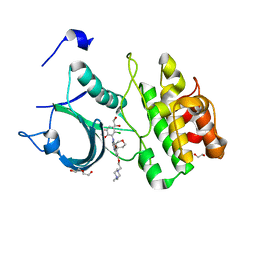 | |
