2P3D
 
 | | Crystal Structure of the multi-drug resistant mutant subtype F HIV protease complexed with TL-3 inhibitor | | Descriptor: | Pol protein, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
2P3A
 
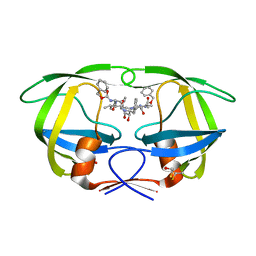 | | Crystal Structure of the multi-drug resistant mutant subtype B HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
1ZVJ
 
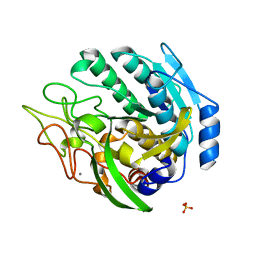 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
6Z2C
 
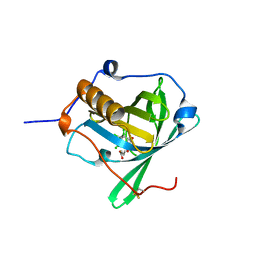 | | Engineered lipocalin C3A5 in complex with a transition state analog | | Descriptor: | 1,7,8,9,10,10-hexachloro-4-carboxypentyl-4-aza-tricyclo[5.2.1.0(2,6)]dec-8-ene-3,5-dione, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an engineered lipocalin that catalyzes a Diels-Alder reaction
To be published
|
|
6Z6Z
 
 | |
2FIV
 
 | | Crystal structure of feline immunodeficiency virus protease complexed with a substrate | | Descriptor: | ACE-ALN-VAL-STA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-21 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
1ZVK
 
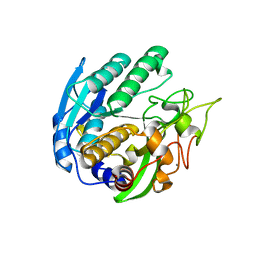 | | Structure of Double mutant, D164N, E78H of Kumamolisin-As | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
3JTK
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055 | | Descriptor: | (2R)-3-benzyl-2-(2-bromo-4-hydroxy-5-methoxyphenyl)-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055
To be Published
|
|
1YG9
 
 | | The structure of mutant (N93Q) of bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aspartic protease Bla g 2, ... | | Authors: | Li, M, Gustchina, A, Wuenschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition.
J.Mol.Biol., 348, 2005
|
|
2P3B
 
 | | Crystal Structure of the subtype B wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
2P3C
 
 | | Crystal Structure of the subtype F wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | ACETIC ACID, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
1Z0C
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain D508A mutant | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0W
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain at 1.2A resolution | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
3KHD
 
 | | Crystal Structure of PFF1300w. | | Descriptor: | Pyruvate kinase | | Authors: | Wernimont, A.K, Hutchinson, A, Hassanali, A, Mackenzie, F, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J.C, Bakszt, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of PFF1300w.
To be Published
|
|
7U9I
 
 | | Co-crystal structure of human CARM1 in complex with MT556 inhibitor | | Descriptor: | 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Perveen, S, Dong, A, Hutchinson, A, Seitova, A, Gibson, E, Hajian, T, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human CARM1 in complex with MT556 inhibitor
To Be Published
|
|
3LIJ
 
 | | Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP | | Descriptor: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, MAGNESIUM ION, ... | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-25 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of full length CpCDPK3 (cgd5_820) in complex with Ca2+ and AMPPNP
To be Published
|
|
1Z0B
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain E506A mutant | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
3LIZ
 
 | | crystal structure of bla g 2 complexed with Fab 4C3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C3 monoclonal antibody Heavy Chain, ... | | Authors: | Li, M, Gustchina, A, Glesner, J, Wunschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding.
Plos One, 6, 2011
|
|
3MA8
 
 | | Crystal structure of CGD1_2040, a pyruvate kinase from cryptosporidium Parvum | | Descriptor: | CITRIC ACID, Pyruvate kinase, SULFATE ION | | Authors: | Wernimont, A.K, Hutchinson, A, Hassanali, A, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of CGD1_2040, a pyruvate kinase from cryptosporidium Parvum
To be Published
|
|
1FMB
 
 | | EIAV PROTEASE COMPLEXED WITH THE INHIBITOR HBY-793 | | Descriptor: | EIAV PROTEASE, [2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYL-VALINYL]-PHENYLALANINOL | | Authors: | Wlodawer, A, Gustchina, A, Zdanov, A, Kervinen, J. | | Deposit date: | 1996-02-27 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of equine infectious anemia virus proteinase complexed with an inhibitor.
Protein Sci., 5, 1996
|
|
1FIV
 
 | | STRUCTURE OF AN INHIBITOR COMPLEX OF PROTEINASE FROM FELINE IMMUNODEFICIENCY VIRUS | | Descriptor: | FIV PROTEASE, FIV PROTEASE INHIBITOR ACE-ALN-VAL-STA-GLU-ALN-NH2 | | Authors: | Wlodawer, A, Gustchina, A, Reshetnikova, L, Lubkowski, J, Zdanov, A. | | Deposit date: | 1995-05-04 | | Release date: | 1995-07-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus.
Nat.Struct.Biol., 2, 1995
|
|
3L19
 
 | | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820 | | Descriptor: | CALCIUM ION, Calcium/calmodulin dependent protein kinase with a kinase domain and 4 calmodulin like EF hands, GLYCEROL, ... | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Walker, J.R, Sullivan, H, Lin, Y.-H, Mackenzie, F, Kozieradzki, I, Cossar, D, Schapira, M, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of calcium binding domain of CpCDPK3, cgd5_820
To be Published
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
1G0V
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
2FMB
 
 | | EIAV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, EQUINE INFECTIOUS ANEMIA VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
