7XAR
 
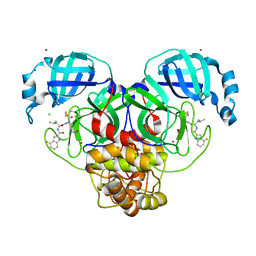 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | Deposit date: | 2022-03-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
2HYM
 
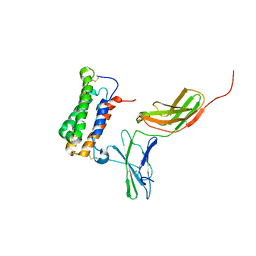 | | NMR based Docking Model of the Complex between the Human Type I Interferon Receptor and Human Interferon alpha-2 | | Descriptor: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | Authors: | Quadt-Akabayov, S.R, Chill, J.H, Levy, R, Kessler, N, Anglister, J. | | Deposit date: | 2006-08-07 | | Release date: | 2006-10-10 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex
Protein Sci., 15, 2006
|
|
2LOX
 
 | |
2JTX
 
 | |
6ZL0
 
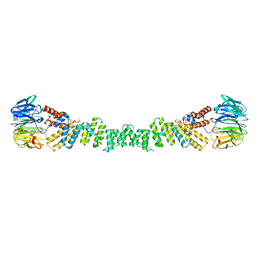 | | COPII on membranes, outer coat left-handed rod | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Hutchings, J. | | Deposit date: | 2020-06-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | Structure of the complete, membrane-assembled COPII coat reveals a complex interaction network.
Nat Commun, 12, 2021
|
|
6ZG5
 
 | |
6T8Y
 
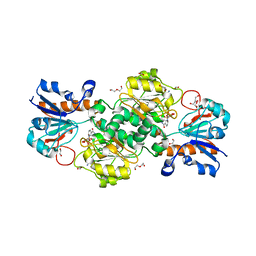 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T94
 
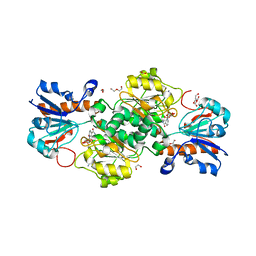 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
2G8G
 
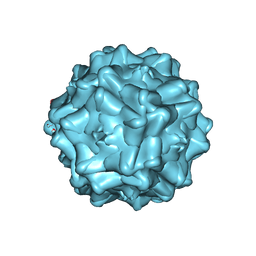 | | Structurally mapping the diverse phenotype of Adeno-Associated Virus serotype 4 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid | | Authors: | Govindasamy, L, Padron, E, McKenna, R, Muzyczka, N, Chiorini, J.A, Agbandje-McKenna, M. | | Deposit date: | 2006-03-02 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally mapping the diverse phenotype of adeno-associated virus serotype 4.
J.Virol., 80, 2006
|
|
6T92
 
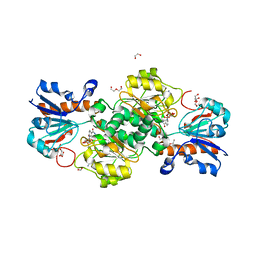 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
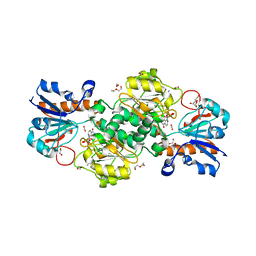 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
1LNS
 
 | |
1CF2
 
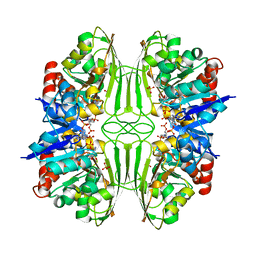 | | THREE-DIMENSIONAL STRUCTURE OF D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC ARCHAEON METHANOTHERMUS FERVIDUS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Charron, C, Talfournier, F, Isuppov, M.N, Branlant, G, Littlechild, J.A, Vitoux, B, Aubry, A. | | Deposit date: | 1999-03-24 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5ZQK
 
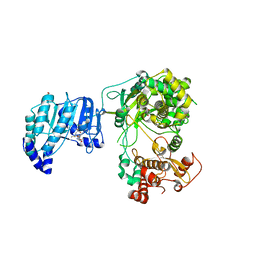 | | Dengue Virus Non Structural Protein 5 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | El Sahili, A, Soh, T.S, Schiltz, J, Gharbi-Ayachi, A, Goh, B.C, Seh, C.C, Dedon, P.C, Shi, P.Y, Lim, S.P, Lescar, J. | | Deposit date: | 2018-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues.
J.Virol., 94, 2019
|
|
2L8B
 
 | | TraI (381-569) | | Descriptor: | Protein traI | | Authors: | Wright, N.T, Raththagala, M.U, Edwards, S, Krueger, S, Schildbach, J.F. | | Deposit date: | 2011-01-07 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and small angle scattering analysis of TraI (381-569).
Proteins, 80, 2012
|
|
1A2Z
 
 | |
8PAJ
 
 | | Crystal Structure of a Squalene-Hopene cyclase from Archangium gephyra | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Worthy, H.L, Isupov, M.N, Littlechild, J.A, Mitchell, D.E. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of a mesophilic Squalene-Hopene Cyclases from Cystobacter fuscus and Archangium gephyra
To Be Published
|
|
8PAK
 
 | | Crystal Structure of a Squalene-Hopene Cyclase from Cystobacter fuscus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Worthy, H.L, Mitchell, D.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of Mesophilic Squalene-Hopene Cyclases from Cystobacter fuscus and Archangium gephyra
To Be Published
|
|
8T31
 
 | | Crystal structure of GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2 | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
5IAE
 
 | | Caspase 3 V266F | | Descriptor: | ACE-ASP-GLU-VAL-ASK, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6W8I
 
 | | Ternary complex structure - BTK cIAP compound 15 | | Descriptor: | 14-{[(3S)-2-(N-methyl-L-alanyl-3-methyl-L-valyl)-3-{[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]carbamoyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}-3,6,9,12-tetraoxatetradecan-1-yl (3R)-3-{5-amino-4-carbamoyl-3-[4-(2,4-difluorophenoxy)phenyl]-1H-pyrazol-1-yl}piperidine-1-carboxylate, Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ... | | Authors: | Calabrese, M.F, Schiemer, J.S. | | Deposit date: | 2020-03-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Characterization of BTK:PROTAC:cIAP Ternary Complexes: From Snapshots to Ensembles
Nat.Chem.Biol., 2020
|
|
7WMP
 
 | | Tail structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Adaptor protein gp12, Nozzle protein gp25, Portal protein | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-01-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori bacteriophage KHP30
To Be Published
|
|
6W7O
 
 | | Ternary complex structure - BTK cIAP compound 17 | | Descriptor: | Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ZINC ION, ... | | Authors: | Calabrese, M.F, Schiemer, J.S. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Characterization of BTK:PROTAC:cIAP Ternary Complexes: From Snapshots to Ensembles
Nat.Chem.Biol., 2020
|
|
6KNR
 
 | | Crystal structure of Estrogen-related receptor gamma ligand-binding domain with DN200699 | | Descriptor: | (E)-4-(1-(4-(1-cyclopropylpiperidin-4-yl)phenyl)-5-hydroxy-2-phenylpent-1-en-1-yl)phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Song, J, Cho, S.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | An orally available inverse agonist of estrogen-related receptor gamma showed expanded efficacy for the radioiodine therapy of poorly differentiated thyroid cancer.
Eur.J.Med.Chem., 205, 2020
|
|
1IYF
 
 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
