6EM5
 
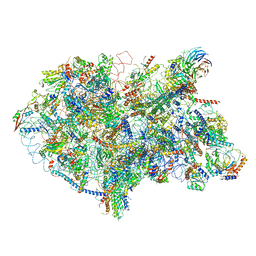 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EZ8
 
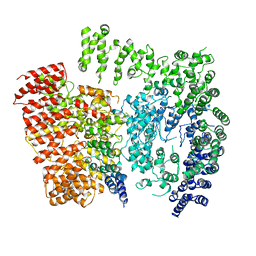 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
6G5I
 
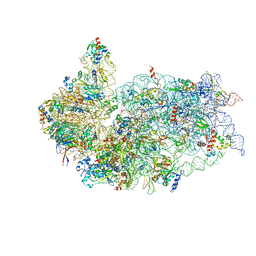 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State R | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G53
 
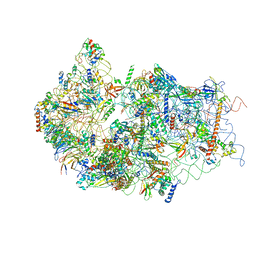 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State E | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G18
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State C | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4S
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State B | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G5H
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - Mature | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4W
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State A | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G51
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State D | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
8JNN
 
 | | linker protein LPP1 from red algal Porphyridium purpureum. | | Descriptor: | LPP1 | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNG
 
 | | Methanesulfonate monooxygenase ferredoxin subunit of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Methanesulfonate monooxygenase ferredoxin subunit | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNL
 
 | | Psb34 from red algal Porphyridium purpureum. | | Descriptor: | Psb34 | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNM
 
 | | PsbW from red algal Porphyridium purpureum. | | Descriptor: | PsbW | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
5LS2
 
 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6T7I
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CGA inhibitory codon combination. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6ZON
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZLW
 
 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6T4Q
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CCG inhibitory codon combination. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6ZN5
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMT
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
7OJ0
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
