3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | Descriptor: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | Authors: | Chen, X, Zhang, Y, Xia, T, Wang, R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
6IG5
 
 | |
6IGA
 
 | |
8WQ3
 
 | | Crystal structure of the C-terminal RRM domain of an RBP | | Descriptor: | CHLORIDE ION, RNA-binding protein 45 | | Authors: | Chen, X, Jiang, M, Yang, Z, Chen, X, Wei, Q, Guo, S, Wang, M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
8WQ5
 
 | | Crystal structure of the C-terminal RRM domain of an RBP in complex with ssDNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*GP*CP*AP*G)-3'), GLYCEROL, RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Chen, X, Wei, Q, Guo, S, Jiang, M, Wang, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
3F9V
 
 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1CN3
 
 | |
8KFO
 
 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bioinspired two-stage assembled photosensitive protein engineering for tumor-specific mitochondrial targeted phototherapy
To Be Published
|
|
7WUH
 
 | | SARS-CoV-2 Spike in complex with Fab of m31A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models.
Sci Transl Med, 14, 2022
|
|
6CIJ
 
 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
8WDD
 
 | | Crystal structure of BSA in complex with B1 | | Descriptor: | Albumin, ~{N},~{N}-dimethyl-6-[(~{E})-2-(1-methylpyridin-1-ium-4-yl)ethenyl]naphthalen-2-amine | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of BSA in complex with B1
To Be Published
|
|
6CG0
 
 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
4JHD
 
 | | Crystal Structure of an Actin Dimer in Complex with the Actin Nucleator Cordon-Bleu | | Descriptor: | Actin-5C, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis of actin filament nucleation by tandem w domains.
Cell Rep, 3, 2013
|
|
7DWS
 
 | |
6DUP
 
 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH COMPOUND 7 | | Descriptor: | (2S)-2-({[3'-(trifluoromethyl)[1,1'-biphenyl]-4-yl]oxy}methyl)-2,3-dihydro-7H-[1,3]oxazolo[3,2-a]pyrimidin-7-one, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Chen, X, Zhang, Y, Mclean, L.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7F3M
 
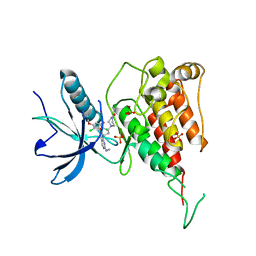 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7F8L
 
 | |
8QH2
 
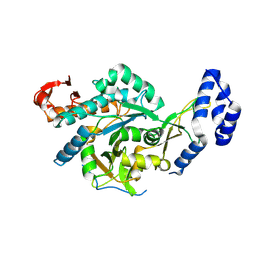 | | Crystal structure of chimeric UAP1L1 | | Descriptor: | UDP-N-acetylhexosamine pyrophosphorylase,UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | | Authors: | Chen, X, Yan, K, Bartual, S, van Aalten, D. | | Deposit date: | 2023-09-06 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | UAP1L1 is an active pyrophosphorylase with an active site disulfide regulating protein thermostability
To Be Published
|
|
8U5H
 
 | |
7WCL
 
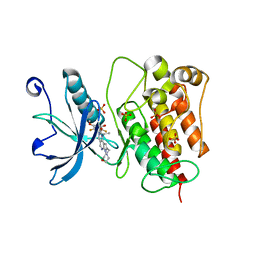 | | Crystal structure of FGFR1 kinase domain with Pemigatinib | | Descriptor: | 11-[2,6-bis(fluoranyl)-3,5-dimethoxy-phenyl]-13-ethyl-4-(morpholin-4-ylmethyl)-5,7,11,13-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),2(6),3,7-tetraen-12-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Lin, Q.M, Jiang, L.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants.
Commun Chem, 5, 2022
|
|
6IHG
 
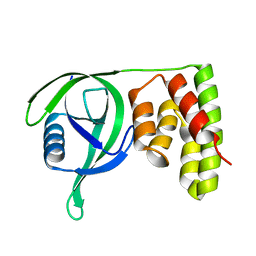 | | N terminal domain of Mycobacterium avium complex Lon protease | | Descriptor: | Lon protease | | Authors: | Chen, X.Y, Zhang, S.J, Bi, F.K, Guo, C.Y, Yao, H.W, Lin, D.H. | | Deposit date: | 2018-09-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
5IRS
 
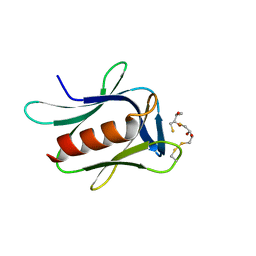 | | crystal structure of the proteasomal Rpn13 PRU-domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Chen, X, Shi, K, Walters, K, Aihara, H. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
2H6O
 
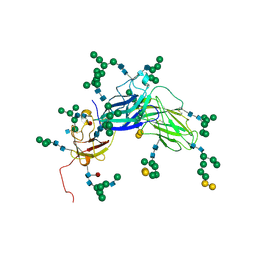 | | Epstein Barr Virus Major Envelope Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major outer envelope glycoprotein gp350, ... | | Authors: | Chen, X.S. | | Deposit date: | 2006-05-31 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Epstein-Barr virus major envelope glycoprotein
Nat.Struct.Mol.Biol., 13, 2006
|
|
5DIK
 
 | | Crystal structure of apo-lpg0406, a carboxymuconolactone decarboxylase family protein from Legionella pneumophila | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Chen, X, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila
Protein Sci., 24, 2015
|
|
