7K0Y
 
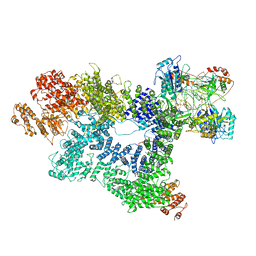 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
6CG0
 
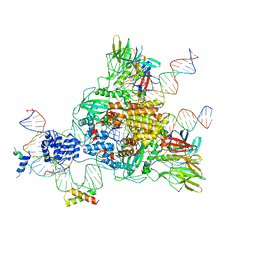 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
7F4G
 
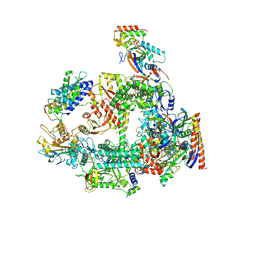 | | Structure of RPAP2-bound RNA polymerase II | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2021-07-07 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
3LUT
 
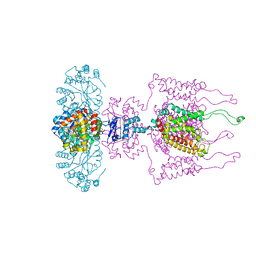 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7CNG
 
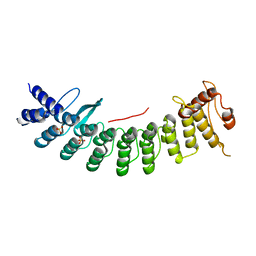 | | Structure of CDK5R1 bound FEM1B | | Descriptor: | Protein fem-1 homolog B,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6DUP
 
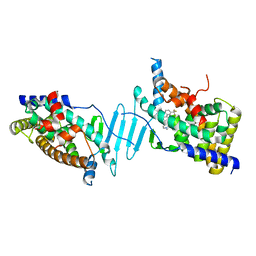 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH COMPOUND 7 | | Descriptor: | (2S)-2-({[3'-(trifluoromethyl)[1,1'-biphenyl]-4-yl]oxy}methyl)-2,3-dihydro-7H-[1,3]oxazolo[3,2-a]pyrimidin-7-one, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Chen, X, Zhang, Y, Mclean, L.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6CIJ
 
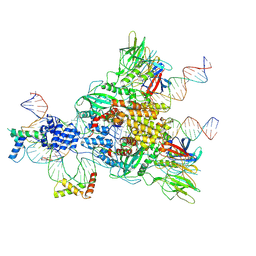 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
3DR9
 
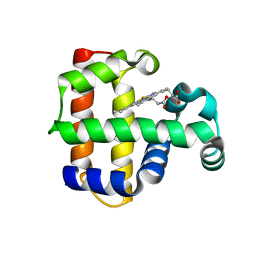 | | Increased Distal Histidine Conformational Flexibility in the Deoxy Form of Dehaloperoxidase from Amphitrite ornata | | Descriptor: | Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Chen, X, de Serrano, V.S, Betts, L, Franzen, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Distal histidine conformational flexibility in dehaloperoxidase from Amphitrite ornata.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EJJ
 
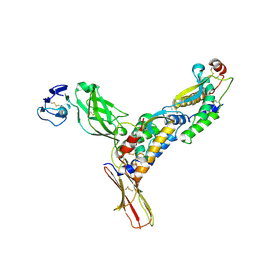 | | Structure of M-CSF bound to the first three domains of FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Colony stimulating factor-1, Macrophage colony-stimulating factor 1 receptor | | Authors: | Chen, X, Liu, H, Focia, P.J, Shim, A, He, X. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6IGA
 
 | |
8XKF
 
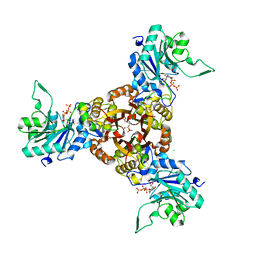 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XHU
 
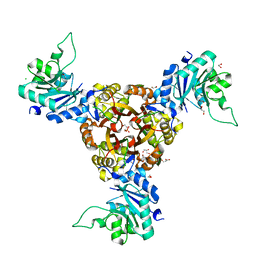 | | Crystal structure of Helicobacter pylori IspDF | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XKG
 
 | | Crystal structure of Acinetobacter baumannii IspD | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, GLYCEROL | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
6IG5
 
 | |
3B3Q
 
 | | Crystal structure of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, X, Liu, H, Shim, A, Focia, P, He, X. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7K11
 
 | |
6JPE
 
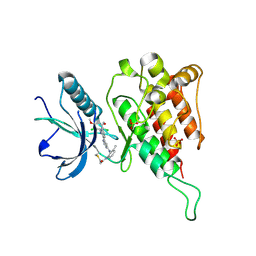 | | Crystal structure of FGFR4 kinase domain with irreversible inhibitor 1 | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[6-[2-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]amino]pyridin-3-yl]pyrimidin-4-yl]amino]-3-methyl-phenyl]prop-2-enamide, SULFATE ION | | Authors: | Chen, X, Dai, S, Zhou, Z, Chen, Y. | | Deposit date: | 2019-03-26 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Development of a Potent and Specific FGFR4 Inhibitor for the Treatment of Hepatocellular Carcinoma.
J.Med.Chem., 63, 2020
|
|
6K9C
 
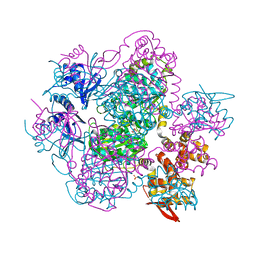 | | The apo structure of NrS-1 C terminal region (305-718) | | Descriptor: | MERCURY (II) ION, Primase, SULFATE ION | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural studies reveal a ring-shaped architecture of deep-sea vent phage NrS-1 polymerase.
Nucleic Acids Res., 48, 2020
|
|
8KFO
 
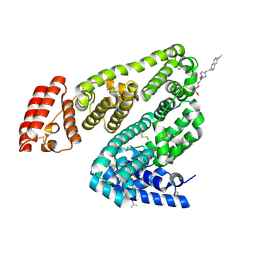 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bioinspired two-stage assembled photosensitive protein engineering for tumor-specific mitochondrial targeted phototherapy
To Be Published
|
|
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Wang, W, Wang, M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
8GI6
 
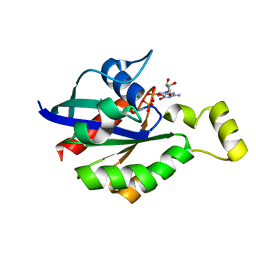 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
6OEO
 
 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
159D
 
 | | SIDE BY SIDE BINDING OF TWO DISTAMYCIN A DRUGS IN THE MINOR GROOVE OF AN ALTERNATING B-DNA DUPLEX | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*IP*CP*IP*CP*IP*CP*IP*C)-3'), MAGNESIUM ION | | Authors: | Chen, X, Ramakrishnan, B, Rao, S.T, Sundaralingam, M. | | Deposit date: | 1994-02-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Nat.Struct.Biol., 1, 1994
|
|
6OEQ
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
