3WRF
 
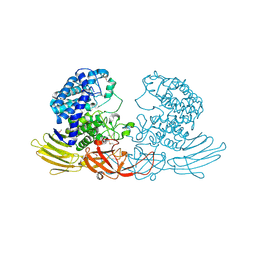 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
5WQD
 
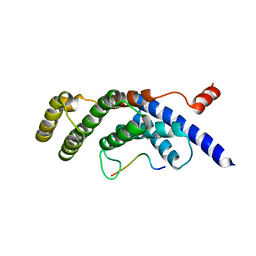 | |
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8D1W
 
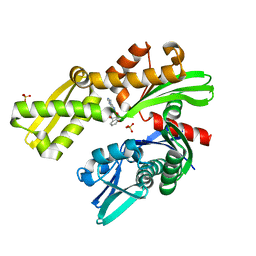 | | Crystal structure of Plasmodium falciparum GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 8-{[(2-chlorophenyl)methyl]amino}adenosine, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W, Smil, D, Zepeda, C.A.V. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Plasmodium falciparum GRP78: nucleoside analogues as agents against the malaria chaperone
Frontiers in Molecular Bioscience, 2022
|
|
6LC1
 
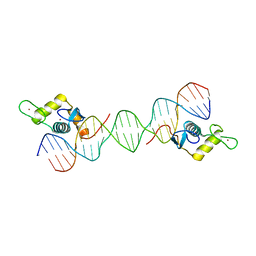 | |
4OI6
 
 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
1IG6
 
 | | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES | | Descriptor: | MODULATOR RECOGNITION FACTOR 2 | | Authors: | Lin, D, Tsui, V, Case, D, Yuan, Y.C, Chen, Y. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES
To be Published
|
|
4OI3
 
 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSP
 
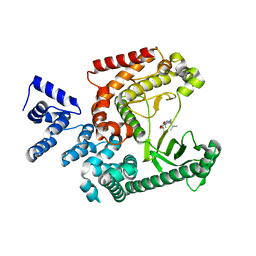 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
8DDI
 
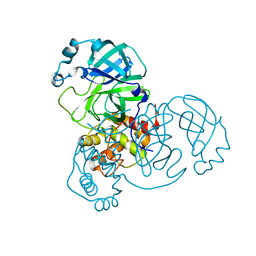 | |
8DDM
 
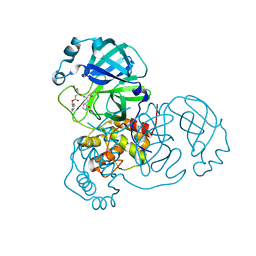 | |
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
3TKL
 
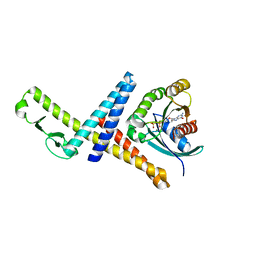 | | Crystal structure of the GTP-bound Rab1a in complex with the coiled-coil domain of LidA from Legionella pneumophila | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, LidA protein, substrate of the Dot/Icm system, ... | | Authors: | Cheng, W, Yin, K, Lu, D, Li, B, Zhu, D, Chen, Y, Zhang, H, Xu, S, Chai, J, Gu, L. | | Deposit date: | 2011-08-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state
Plos Pathog., 8, 2012
|
|
2QPE
 
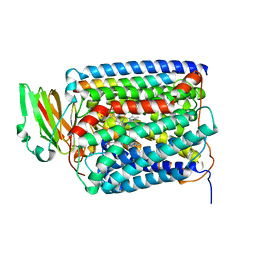 | | An unexpected outcome of surface-engineering an integral membrane protein: Improved crystallization of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Luna, V.M, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba(3) from Thermus thermophilus.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2QPD
 
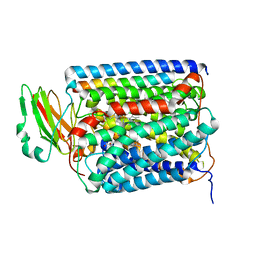 | | An unexpected outcome of surface-engineering an integral membrane protein: Improved crystallization of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Luna, V.M, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba(3) from Thermus thermophilus.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4HMP
 
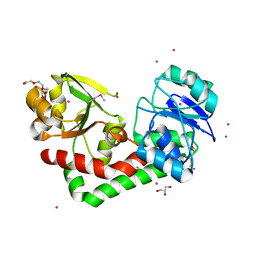 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
4HMO
 
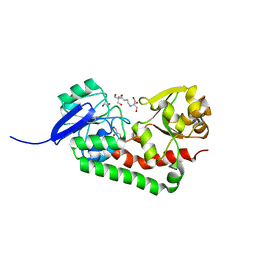 | | Crystal structure of streptococcus pneumoniae TIGR4 PiaA in complex with Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Iron-compound ABC transporter, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
4HMQ
 
 | | Crystal structure of streptococcus pneumoniae TIGR4 PiaA in complex with ferrichrome | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
