9K36
 
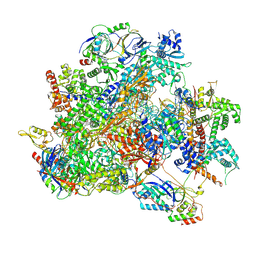 | | Human RNA Polymerase III de novo transcribing complex 12 (TC12) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (54-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | human RNA polymerase III de novo transcribing complex 12
To Be Published
|
|
9LXO
 
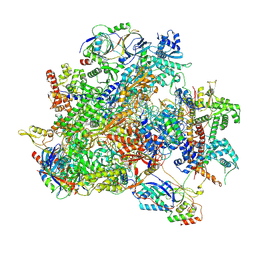 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human RNA Polymerase III de novo transcribing complex 5 (TC5)(without 3'dATP)
To Be Published
|
|
9LXN
 
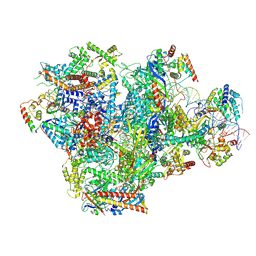 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Human RNA Polymerase III de novo transcribing complex 5 (TC5)(without 3'dATP)
To Be Published
|
|
9K3U
 
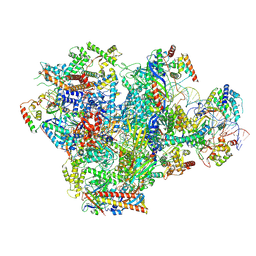 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5) | | Descriptor: | DNA (97-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | human RNA polymerase III de novo transcribing complex 4
To Be Published
|
|
9K2G
 
 | | Human RNA Polymerase III de novo transcribing complex 13 (TC13) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (104-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-17 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human RNA Polymerase III de novo transcribing complex 14 (TC14)
To Be Published
|
|
9K38
 
 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (51-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human RNA Polymerase III de novo transcribing complex 6 (TC6)
To Be Published
|
|
9K39
 
 | | Human RNA Polymerase III de novo transcribing complex 8 (TC8) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (50-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Human RNA Polymerase III de novo transcribing complex 8 (TC8)
To Be Published
|
|
9K3V
 
 | | Human RNA Polymerase III de novo transcribing complex 4 (TC4) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (96-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-20 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Human RNA Polymerase III de novo transcribing complex 4 (TC4)
To Be Published
|
|
9K3B
 
 | | Human RNA Polymerase III de novo transcribing complex 10 overall (TC10-overall) | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA (102-MER), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2024-10-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | human RNA polymerase III de novo transcribing complex 10-overall
To Be Published
|
|
7YCX
 
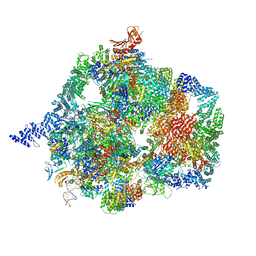 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1HKA
 
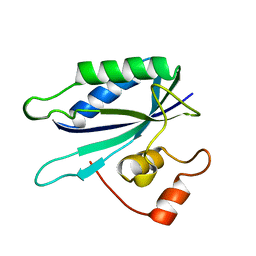 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
4GHQ
 
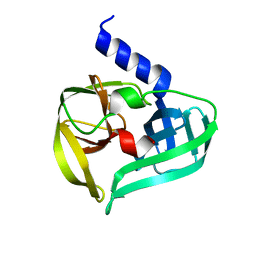 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
9JVP
 
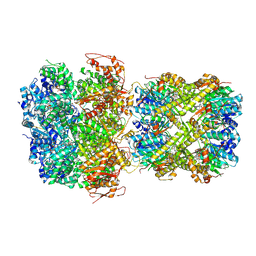 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
8Z1E
 
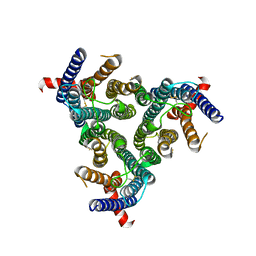 | | A homotrimeric GPCR architecture of the human cytomegalovirus (UL78) revealed by cryo-EM | | Descriptor: | Uncharacterized protein UL78 | | Authors: | Chen, Y, Li, Y, Zhou, Q, Cong, Z, Lin, S, Yan, J, Chen, X, Yang, D, Ying, T, Wang, M.-W. | | Deposit date: | 2024-04-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A homotrimeric GPCR architecture of the human cytomegalovirus revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8XU7
 
 | | Crystal structure of Lsd18 in complex with a product | | Descriptor: | (4R,5S)-3-((2R,3S,4S)-2-ethyl-5-((2R,3R)-2-ethyl-3-(2-((2R,3R)-2-ethyl-3-methyloxiran-2-yl)ethyl)oxiran-2-yl)-3-hydroxy-4-methylpentanoyl)-4-methyl-5-phenyloxazolidin-2-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Deng, Y.M, Chen, X. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Sequential Enantioselective Epoxidation by a Flavin-Dependent Monooxygenase in Lasalocid A Biosynthesis.
Angew.Chem.Int.Ed.Engl., 2025
|
|
1WWN
 
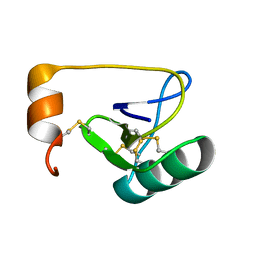 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
8XTZ
 
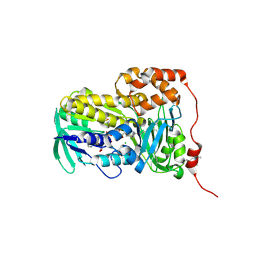 | | Crystal structure of Lsd18 in complex with a substrate | | Descriptor: | (4R,5S)-4-methyl-5-phenyl-3-((2R,3S,4S,6E,10E)-2,6,10-triethyl-3-hydroxy-4-methyldodeca-6,10-dienoyl)oxazolidin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Deng, Y.M, Hu, Y.L, Chen, X. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Sequential Enantioselective Epoxidation by a Flavin-Dependent Monooxygenase in Lasalocid A Biosynthesis.
Angew.Chem.Int.Ed.Engl., 2025
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
8YRP
 
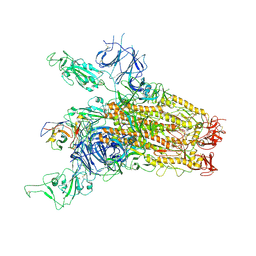 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRO
 
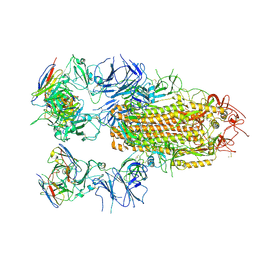 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
7QZQ
 
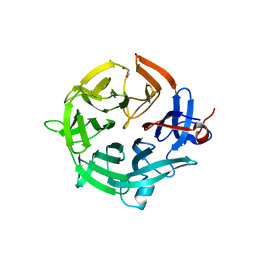 | | Crystal structure of the kelch domain of human KBTBD12 | | Descriptor: | 1,2-ETHANEDIOL, Kelch repeat and BTB domain-containing protein 12, SODIUM ION | | Authors: | Manning, C.E, Chen, Z, Chen, X, Bradshaw, W.J, Bakshi, S, Mckinley, G, Chalk, R, Burgess-Brown, N, von Delft, F, Bullock, A.N. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the kelch domain of human KBTBD12
To Be Published
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
8Z98
 
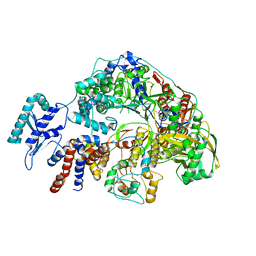 | | Cryo-EM structure of Thogoto virus polymerase in a transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z90
 
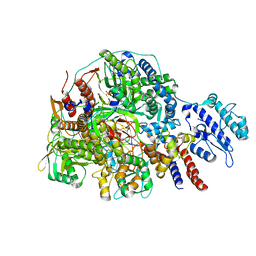 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
