4JD6
 
 | | Crystal structure of Mycobacterium tuberculosis Eis in complex with coenzyme A and tobramycin | | Descriptor: | COENZYME A, Enhanced intracellular survival protein, TOBRAMYCIN | | Authors: | Biswas, T, Chen, W, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2013-02-23 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase eis.
Chembiochem, 14, 2013
|
|
4DNW
 
 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4P5B
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
8XC4
 
 | | Nipah virus attachment glycoprotein head domain in complex with a broadly neutralizing antibody 1E5 | | Descriptor: | 1E5-VH, 1E5-VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, P.F, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A potent Henipavirus cross-neutralizing antibody reveals a dynamic fusion-triggering pattern of the G-tetramer.
Nat Commun, 15, 2024
|
|
6IZJ
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6K2H
 
 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
8JA5
 
 | | Crystal structure of Nipah Virus attachment (G) glycoprotein in complex with neutralizing antibody 14F8 | | Descriptor: | 14F8 antibody heavy chain, 14F8 antibody light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Y.H, Huang, X.Y, Xu, J.J, Chen, W. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Nipah Virus attachment (G) glycoprotein in complex with neutralizing antibody 14F8
To Be Published
|
|
8JR5
 
 | |
8JR3
 
 | | Crystal structure of Hendra Virus attachment(G) glycoprotein mutant S586N in complex with neutralizing antibody 14F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Y.H, Huang, X.Y, Xu, J.J, Chen, W. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal structure of Hendra Virus attachment
(G) glycoprotein mutant S586N in complex with neutralizing antibody 14F8
To Be Published
|
|
8JOV
 
 | | Portal-tail complex of phage GP4 | | Descriptor: | Portal protein, Putative tail fiber protein, Virion associated protein, ... | | Authors: | Liu, H, Chen, W. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
8JOU
 
 | | Fiber I and fiber-tail-adaptor of phage GP4 | | Descriptor: | Virion-associated phage protein, rope protein of phage GP4 | | Authors: | Liu, H, Chen, W. | | Deposit date: | 2023-06-08 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structure of Podophage GP4 Reveals a Novel Architecture of Three Types of Tail Fibers.
J.Mol.Biol., 435, 2023
|
|
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
5IPK
 
 | | Structure of the R432A variant of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
6M0W
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | Descriptor: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0X
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0V
 
 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-22 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
5ZRX
 
 | | Crystal Structure of EphA2/SHIP2 Complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2,Ephrin type-A receptor 2 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5ZRY
 
 | | Crystal Structure of EphA6/Odin Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ankyrin repeat and SAM domain-containing protein 1A,Ephrin type-A receptor 6, ... | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5ZRZ
 
 | | Crystal Structure of EphA5/SAMD5 Complex | | Descriptor: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
8IDE
 
 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | Descriptor: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | Authors: | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
8HUD
 
 | | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target DNA strand, Target DNA strand, ... | | Authors: | Tang, N, Wu, Z, Gao, Y, Chen, W, Su, M, Wang, Z, Ji, Q. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Molecular Basis and Genome Editing Applications of a Compact Eubacterium ventriosum CRISPR-Cas9 System.
Acs Synth Biol, 13, 2024
|
|
5YCO
 
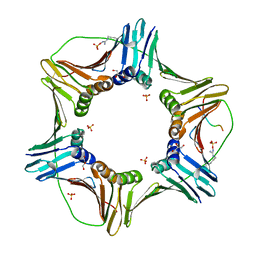 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
