6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1E
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
1CXX
 
 | | MUTANT R122A OF QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | CYSTEINE AND GLYCINE-RICH PROTEIN CRP2, ZINC ION | | Authors: | Kloiber, K, Weiskirchen, R, Kraeutler, B, Bister, K, Konrat, R. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutational analysis and NMR spectroscopy of quail cysteine and glycine-rich protein CRP2 reveal an intrinsic segmental flexibility of LIM domains.
J.Mol.Biol., 292, 1999
|
|
1A7I
 
 | | AMINO-TERMINAL LIM DOMAIN FROM QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | QCRP2 (LIM1), ZINC ION | | Authors: | Kontaxis, G, Konrat, R, Kraeutler, B, Weiskirchen, R, Bister, K. | | Deposit date: | 1998-03-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and intramodular dynamics of the amino-terminal LIM domain from quail cysteine- and glycine-rich protein CRP2.
Biochemistry, 37, 1998
|
|
1J09
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP and Glu | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTAMIC ACID, Glutamyl-tRNA synthetase, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
4K5Y
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, ... | | Authors: | Hollenstein, K, Kean, J, Bortolato, A, Cheng, R.K.Y, Dore, A.S, Jazayeri, A, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2013-04-15 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.977 Å) | | Cite: | Structure of class B GPCR corticotropin-releasing factor receptor 1.
Nature, 499, 2013
|
|
1HCW
 
 | |
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
1QLI
 
 | | QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYSTEINE AND GLYCINE-RICH PROTEIN, ZINC ION | | Authors: | Konrat, R, Weiskirchen, R, Krautler, B, Bister, K. | | Deposit date: | 1997-02-17 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carboxyl-terminal LIM domain from quail cysteine-rich protein CRP2.
J.Biol.Chem., 272, 1997
|
|
2ZZQ
 
 | | Crystal structure analysis of the HEV capsid protein, PORF2 | | Descriptor: | Protein ORF3, Capsid protein | | Authors: | Miyazaki, N, Xing, L, Wang, C.-Y, Li, T.-C, Takeda, N, Higashiura, A, Nakagawa, A, Tsukihara, T, Miyamura, T, Cheng, R.H. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Role of protein domain-modularity in designating capsid assembly and antigenicity
To be Published
|
|
1N75
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N78
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and glutamol-AMP. | | Descriptor: | GLUTAMOL-AMP, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N77
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HZ1
 
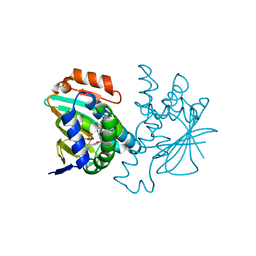 | | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03
TO BE PUBLISHED
|
|
3HYZ
 
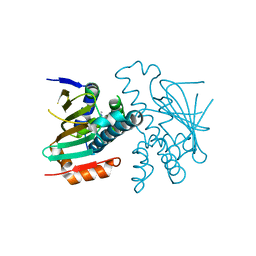 | | Crystal structure of Hsp90 with fragment 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragment 42-C03
TO BE PUBLISHED
|
|
3HZ5
 
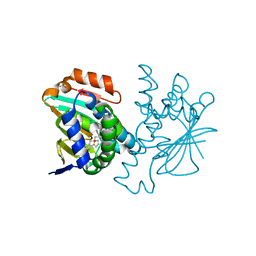 | | Crystal structure of Hsp90 with fragment Z064 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(5-furan-2-yl-3-methyl-1H-pyrazol-4-yl)butyl]-N-methyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment Z064
to be published
|
|
3HOF
 
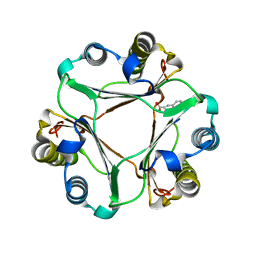 | | Structure of macrophage migration inhibitory factor (MIF) with caffeic acid at 1.9A resolution | | Descriptor: | CAFFEIC ACID, Macrophage migration inhibitory factor | | Authors: | Crawley, L, Barker, J, Cheng, R.K.Y, Wood, M, Felicetti, B. | | Deposit date: | 2009-06-02 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of macrophage migration inhibitory factor (MIF) with caffeic acid at 1.9A resolution
To be Published
|
|
3HYY
 
 | | Crystal structure of Hsp90 with fragment 37-D04 | | Descriptor: | Heat shock protein HSP 90-alpha, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment 37-D04
TO BE PUBLISHED
|
|
3K4S
 
 | | The structure of the catalytic domain of human PDE4d with 4-(3-Butoxy-4-methoxyphenyl)methyl-2-imidazolidone | | Descriptor: | (4R)-4-(3-butoxy-4-methoxybenzyl)imidazolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Crawley, L, Cheng, R.K.Y, Wood, M, Barker, J, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-10-06 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the catalytic domain of human PDE4d with 4-(3-Butoxy-4-methoxyphenyl)methyl-2-imidazolidone
To be Published
|
|
3K5E
 
 | | The structure of human kinesin-like motor protein Kif11/KSP/Eg5 in complex with ADP and enastrol. | | Descriptor: | (4S,5R)-5-hydroxy-4-(3-hydroxyphenyl)-3,4,5,6,7,8-hexahydroquinazoline-2(1H)-thione, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Crawley, L, Cheng, R.K.Y, Wood, M, Barker, J, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of human kinesin-like motor protein Kif11/KSP/Eg5 in complex with ADP and enastrol.
To be Published
|
|
3IYS
 
 | | Homology model of avian polyomavirus asymmetric unit | | Descriptor: | Major capsid protein VP1 | | Authors: | Shen, P.S, Enderlein, D, Nelson, C.D.S, Carter, W.S, Kawano, M, Xing, L, Swenson, R.D, Olson, N.H, Baker, T.S, Cheng, R.H, Atwood, W.J, Johne, R, Belnap, D.M. | | Deposit date: | 2010-04-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | The structure of avian polyomavirus reveals variably sized capsids, non-conserved inter-capsomere interactions, and a possible location of the minor capsid protein VP4.
Virology, 411, 2011
|
|
3FT8
 
 | | Structure of HSP90 bound with a noval fragment. | | Descriptor: | (5E,7S)-2-amino-7-(4-fluoro-2-pyridin-3-ylphenyl)-4-methyl-7,8-dihydroquinazolin-5(6H)-one oxime, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based identification of Hsp90 inhibitors.
Chemmedchem, 4, 2009
|
|
