8ZE0
 
 | |
8ZE2
 
 | |
8ZMN
 
 | | Crystal structure of ANTXR1 | | Descriptor: | Anthrax toxin receptor 1, SODIUM ION | | Authors: | Zheng, H, Hu, J, Fu, L, Chen, R. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structure of Anthrax toxin receptor 1 APO from Rattus norvegicus
To Be Published
|
|
6L2A
 
 | | A mutant form of M. tb toxin MazEF-mt1 | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.90044665 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6KYS
 
 | | The structure of the M. tb toxin MazF-mt1 | | Descriptor: | Endoribonuclease MazF9 | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-09-20 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200414 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6WPS
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WS6
 
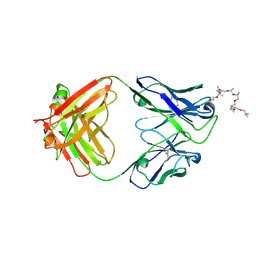 | | Structural and functional analysis of a potent sarbecovirus neutralizing antibody | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), S309 antigen-binding (Fab) fragment, heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, Marco, A.D, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Telenti, A, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
2VVW
 
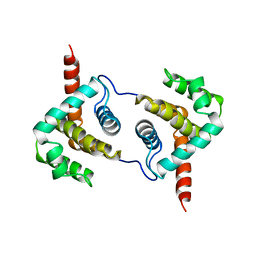 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
6WPT
 
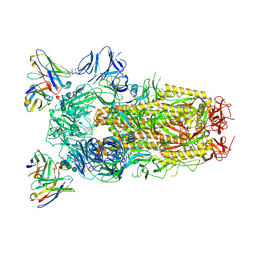 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
2VVY
 
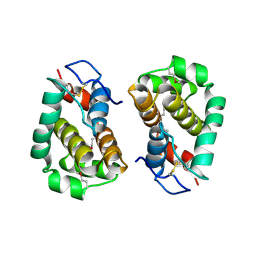 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VVX
 
 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2UXE
 
 | | The structure of Vaccinia virus N1 | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Cooray, S, Bahar, M.W, Abrescia, N.G.A, McVey, C.E, Bartlett, N.W, Chen, R.A.-J, Stuart, D.I, Grimes, J.M, Smith, G.L. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and Structural Studies of the Vaccinia Virus Virulence Factor N1 Reveal a Bcl-2-Like Anti- Apoptotic Protein
J.Gen.Virol., 88, 2007
|
|
1U4D
 
 | | Structure of the ACK1 Kinase Domain bound to Debromohymenialdisine | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, DEBROMOHYMENIALDISINE | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
1U54
 
 | | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the CDC42-associated Tyrosine Kinase ACK1 bound to AMP-PCP | | Descriptor: | Activated CDC42 kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
4ME2
 
 | | Crystal Structure of THA8 protein from Brachypodium distachyon | | Descriptor: | Uncharacterized protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Melcher, K, Xu, H.E. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1U46
 
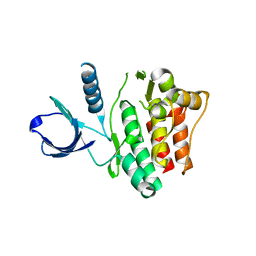 | | Crystal Structure of the Unphosphorylated Kinase Domain of the Tyrosine Kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
4N2Q
 
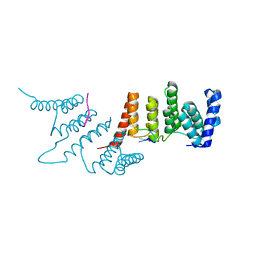 | | Crystal structure of THA8 in complex with Zm4 RNA | | Descriptor: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4N2S
 
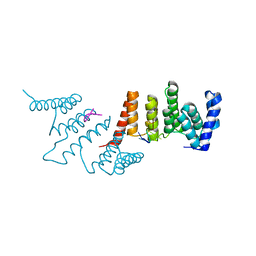 | | Crystal Structure of THA8 in complex with Zm1a-6 RNA | | Descriptor: | THA8 RNA binding protein, Zm1a-6 RNA | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4IH1
 
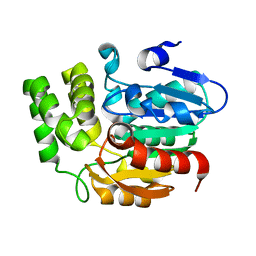 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IHA
 
 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4ZRS
 
 | | Crystal structure of a cloned feruloyl esterase from a soil metagenomic library | | Descriptor: | Esterase, GLYCEROL | | Authors: | Xie, W, Chen, R, Cao, L, Liu, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing the Thermostability of Feruloyl Esterase EstF27 by Directed Evolution and the Underlying Structural Basis
J.Agric.Food Chem., 63, 2015
|
|
3MXX
 
 | |
3N0D
 
 | |
4LEU
 
 | | Crystal Structure of THA8-like protein from Arabidopsis thaliana | | Descriptor: | Pentatricopeptide repeat-containing protein At3g46870 | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Kang, Y, Melcher, K, Zhu, J.K, Xu, H.E. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a PLS-class Pentatricopeptide Repeat Protein Provides Insights into Mechanism of RNA Recognition.
J.Biol.Chem., 288, 2013
|
|
