1D3W
 
 | | Crystal structure of ferredoxin 1 d15e mutant from azotobacter vinelandii at 1.7 angstrom resolution. | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | 著者 | Chen, K, Hirst, J, Camba, R, Bonagura, C.A, Stout, C.D, Burges, B.K, Armstrong, F.A. | | 登録日 | 1999-10-01 | | 公開日 | 1999-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Atomically defined mechanism for proton transfer to a buried redox centre in a protein.
Nature, 405, 2000
|
|
1F5C
 
 | | CRYSTAL STRUCTURE OF F25H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER, ... | | 著者 | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | 登録日 | 2000-06-13 | | 公開日 | 2000-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
1F5B
 
 | | CRYSTAL STRUCTURE OF F2H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | 著者 | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | 登録日 | 2000-06-13 | | 公開日 | 2000-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
5AF0
 
 | | MAEL domain from Bombyx mori Maelstrom | | 分子名称: | MAELSTROM, ZINC ION | | 著者 | Chen, K, Campbell, E, Pandey, R.R, Yang, Z, McCarthy, A.A, Pillai, R.S. | | 登録日 | 2015-01-13 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Metazoan Maelstrom is an RNA-Binding Protein that Has Evolved from an Ancient Nuclease Active in Protists.
RNA, 21, 2015
|
|
2JYW
 
 | | Solution structure of C-terminal domain of APOBEC3G | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | 著者 | Chen, K, Harjes, E, Gross, P.J, Fahmy, A, Lu, Y, Shindo, K, Harris, R.S, Matsuo, H. | | 登録日 | 2007-12-20 | | 公開日 | 2008-02-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
Nature, 452, 2008
|
|
8XVV
 
 | | The TRRAP module of human NuA4/TIP60 complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, Isoform 2 of E1A-binding protein p400, Isoform 2 of Transformation/transcription domain-associated protein | | 著者 | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human TIP60 complex
To Be Published
|
|
8XVT
 
 | | The core subcomplex of human NuA4/TIP60 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human TIP60 complex
To Be Published
|
|
8XVG
 
 | | Structure of human NuA4/TIP60 complex | | 分子名称: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | 著者 | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Structure of the human TIP60 complex
To Be Published
|
|
5Y4F
 
 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | 分子名称: | ACETATE ION, Ankyrin-2, CALCIUM ION | | 著者 | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
8JRU
 
 | | Cryo-EM structure of the glucagon receptor bound to beta-arrestin 1 in ligand-free state | | 分子名称: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, Nanobody 32, ... | | 著者 | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | 登録日 | 2023-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JRV
 
 | | Cryo-EM structure of the glucagon receptor bound to glucagon and beta-arrestin 1 | | 分子名称: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), Glucagon, HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, ... | | 著者 | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | 登録日 | 2023-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
5Y4E
 
 | | Crystal Structure of AnkB Ankyrin Repeats R8-14 in complex with autoinhibition segment AI-b | | 分子名称: | Ankyrin-2,Ankyrin-2, GLYCEROL, SULFATE ION | | 著者 | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.341 Å) | | 主引用文献 | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4D
 
 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | 分子名称: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | 著者 | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
6FWS
 
 | |
6FWR
 
 | | Structure of DinG in complex with ssDNA | | 分子名称: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | 著者 | Cheng, K, Wigley, D.B. | | 登録日 | 2018-03-07 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
8IOO
 
 | |
8IU7
 
 | |
6SJG
 
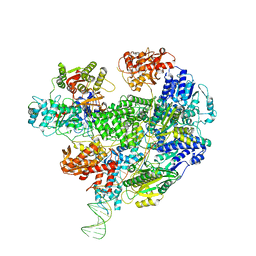 | | Cryo-EM structure of the RecBCD no Chi negative control complex | | 分子名称: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJE
 
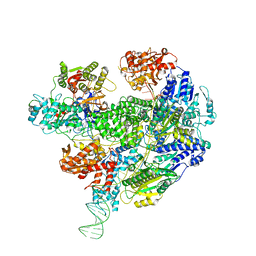 | | Cryo-EM structure of the RecBCD Chi partially-recognised complex | | 分子名称: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T2V
 
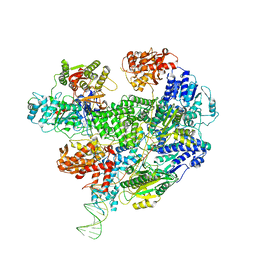 | | Cryo-EM structure of the RecBCD in complex with Chi-plus2 substrate | | 分子名称: | DNA (Chi-plus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-10-09 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJB
 
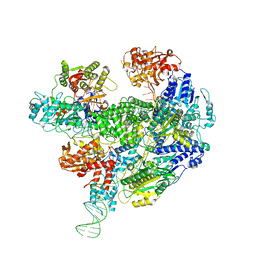 | | Cryo-EM structure of the RecBCD Chi recognised complex | | 分子名称: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T2U
 
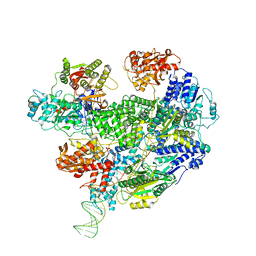 | | Cryo-EM structure of the RecBCD in complex with Chi-minus2 substrate | | 分子名称: | DNA (Chi-minus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-10-09 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SER
 
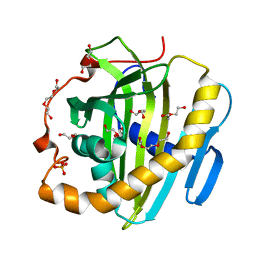 | | Crystal structure of human STARD10 | | 分子名称: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, START domain-containing protein 10, ... | | 著者 | Cheng, K, Wigley, D.B. | | 登録日 | 2019-07-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | The crystal structure of human STARD10
To Be Published
|
|
6SJF
 
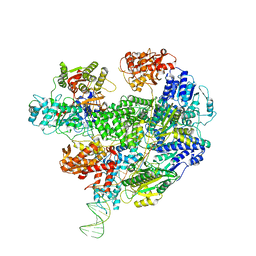 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | 分子名称: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7W8D
 
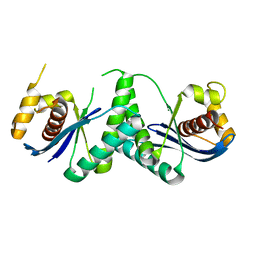 | | The structure of Deinococcus radiodurans RuvC | | 分子名称: | Crossover junction endodeoxyribonuclease RuvC, MAGNESIUM ION | | 著者 | Cheng, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75016141 Å) | | 主引用文献 | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
