4H00
 
 | | The crystal structure of mon-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4QPD
 
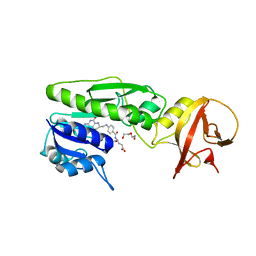 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4GZ7
 
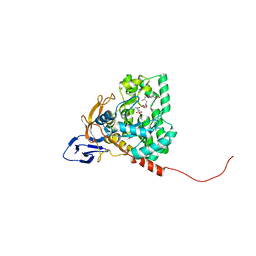 | | The crystal structure of Apo-dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, dihydropyrimidinase | | Authors: | Hsien, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structure Requirements for Post-translational Modification
To be Published
|
|
4H01
 
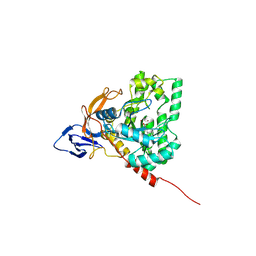 | | The crystal structure of di-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4R8V
 
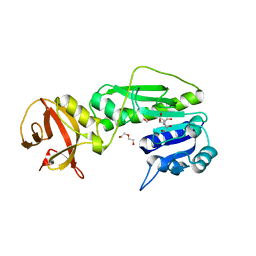 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8W4L
 
 | | Crystal structure of closed conformation of human immunoglobulin Fc in presence of EndoSz | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4M
 
 | | Crystal structure of open conformation of human immunoglobulin Fc in presence of EndoSz | | Descriptor: | Immunoglobulin gamma-1 heavy chain, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4G
 
 | | Crystal structure of EndoSz mutant D234M, from Streptococcus equi subsp. Zooepidemicus Sz105 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4N
 
 | | Crystal structure of EndoSz mutant D234M, in space group P21, in complex with oligosaccharide G2S1 | | Descriptor: | CALCIUM ION, Glycoside hydrolase, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4I
 
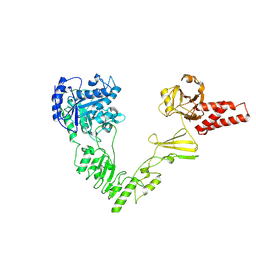 | | Crystal structure of EndoSz mutant D234M in space group P21 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8X8G
 
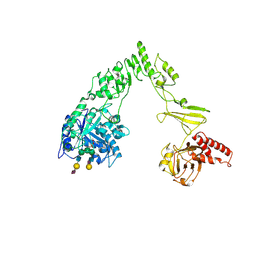 | | Crystal structure of EndoSz mutant D234M, from Streptococcus equi subsp. Zooepidemicus Sz105, in complex with oligosaccharide G2S2-oxazoline | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, CALCIUM ION, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose, ... | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
5XMJ
 
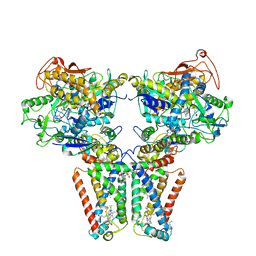 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
5YL0
 
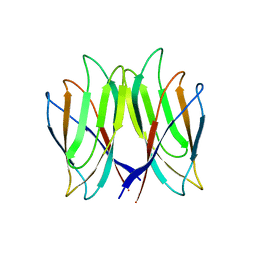 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P212121) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKX
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Cd ion | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, Capsid protein, ... | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKZ
 
 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P21) | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKV
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKU
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Zn ions | | Descriptor: | Capsid protein, ZINC ION | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YL1
 
 | | T=1 subviral particle of Penaeus vannamei nodavirus capsid protein deletion mutant (delta 1-37 & 251-368) | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AB6
 
 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AB5
 
 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
3DC0
 
 | | Crystal structure of native alpha-amylase from Bacillus sp. KR-8104 | | Descriptor: | CALCIUM ION, alpha-amylase | | Authors: | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, M, Naderi-Manesh, H, Chen, C.J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-06-17 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of native alpha-amylase from Bacillus sp. KR-8104.
to be published
|
|
3E5R
 
 | |
3E6A
 
 | |
3K7N
 
 | | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guan, H.H, Huang, Y.W, Wu, W.G, Chen, C.J. | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins
J.Struct.Biol., 169, 2010
|
|
3K7L
 
 | | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Atragin, CALCIUM ION, ... | | Authors: | Guan, H.H, Wu, W.G, Chen, C.J. | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins
J.Struct.Biol., 169, 2010
|
|
