4GIT
 
 | |
6KZW
 
 | | Crystal structure of YggS family pyridoxal phosphate-dependent enzyme PipY from Fusobacterium nucleatum | | Descriptor: | PHOSPHATE ION, Pyridoxal phosphate homeostasis protein | | Authors: | Chen, Y, Wang, L, Shang, F, Lan, J, Liu, W, Xu, Y. | | Deposit date: | 2019-09-25 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of YggS family pyridoxal phosphate-dependent enzyme PipY from Fusobacterium nucleatum
To Be Published
|
|
5X8F
 
 | |
1G6M
 
 | |
1CQ4
 
 | | CI2 MUTANT WITH TETRAGLUTAMINE (MGQQQQGM) REPLACING MET59 | | Descriptor: | PROTEIN (SERINE PROTEINASE INHIBITOR 2), SULFATE ION | | Authors: | Chen, Y.W, Stott, K.R. | | Deposit date: | 1998-11-17 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a dimeric chymotrypsin inhibitor 2 mutant containing an inserted glutamine repeat.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3ZH2
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase in complex with a DNA aptamer | | Descriptor: | DNA APTAMER, L-LACTATE DEHYDROGENASE | | Authors: | Cheung, Y.W, Kwok, J, Law, A.W.L, Watt, R.M, Kotaka, M, Tanner, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Y26
 
 | | Crystal structure of native Dpb4-Dpb3 | | Descriptor: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | Authors: | Chen, Y.H, Li, Y, Gao, F. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1ETO
 
 | |
5YC2
 
 | |
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
1ETQ
 
 | |
1ETV
 
 | |
5YCA
 
 | | Crystal structure of inner membrane protein Bqt4 in complex with LEM2 | | Descriptor: | Lap-Emerin-Man domain protein 2, Ubiquitin-like protein SMT3,Bouquet formation protein 4 | | Authors: | Chen, Y, Hu, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into chromosome attachment to the nuclear envelope by an inner nuclear membrane protein Bqt4 in fission yeast.
Nucleic Acids Res., 47, 2019
|
|
7UL1
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
6A6W
 
 | |
7D1G
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION | | Authors: | Chen, Y, Lan, J, Liu, W, Wang, L, Xu, Y. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii
To Be Published
|
|
5ZB5
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
1ETK
 
 | |
1ETY
 
 | |
1ETX
 
 | |
1ETW
 
 | |
6ALJ
 
 | | ALDH1A2 liganded with NAD and compound WIN18,446 | | Descriptor: | Aldehyde dehydrogenase 1A2, N,N'-(octane-1,8-diyl)bis(2,2-dichloroacetamide), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, Y, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of ALDH1A2 Inhibition by Irreversible and Reversible Small Molecule Inhibitors.
ACS Chem. Biol., 13, 2018
|
|
8TGF
 
 | | Crystal structure of cEPG5 LIR/LGG-1 complex | | Descriptor: | ACETATE ION, Ectopic P granules protein 5, Protein lgg-1 | | Authors: | Cheung, Y.W.S, Yip, C.K. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of the mode of interaction between the tandem LC3-interacting region motif of the autophagy tethering factor EPG5 and ATG8 proteins
To be published
|
|
1SUV
 
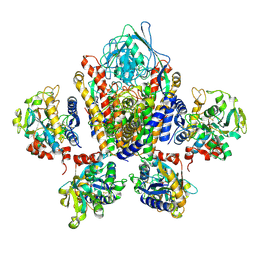 | | Structure of Human Transferrin Receptor-Transferrin Complex | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin, ... | | Authors: | Cheng, Y, Zak, O, Aisen, P, Harrison, S.C, Walz, T. | | Deposit date: | 2004-03-26 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the Human Transferrin Receptor-Transferrin Complex
Cell(Cambridge,Mass.), 116, 2004
|
|
