8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
2GIF
 
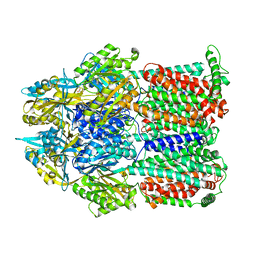 | | Asymmetric structure of trimeric AcrB from Escherichia coli | | Descriptor: | Acriflavine resistance protein B, CITRATE ANION | | Authors: | Seeger, M.A, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K, Pos, K.M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism.
Science, 313, 2006
|
|
6WGT
 
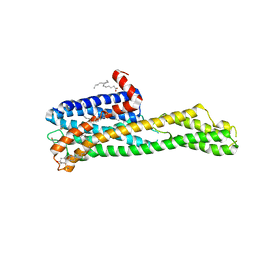 | | Crystal structure of HTR2A with hallucinogenic agonist | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562 fusion, CHOLESTEROL, ... | | Authors: | Kim, K.L, Che, T, Krumm, B.E, Roth, B.L. | | Deposit date: | 2020-04-06 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT 2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6WH4
 
 | | Crystal structure of HTR2A with inverse agonist | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562 fusion, CHOLESTEROL, ... | | Authors: | Kim, K.L, Che, T, Krumm, B.E, Roth, B.L. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT 2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
8DZP
 
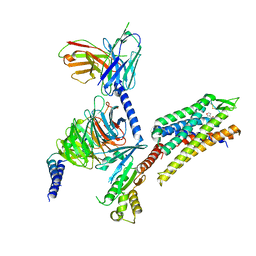 | | momSalB bound Kappa Opioid Receptor in complex with Gi1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZQ
 
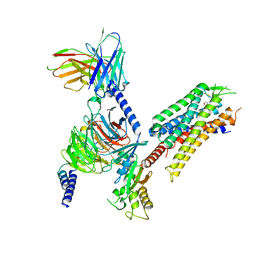 | | momSalB bound Kappa Opioid Receptor in complex with GoA | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZS
 
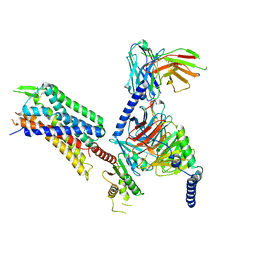 | | GR89,696 bound Kappa Opioid Receptor in complex with Gz | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(z) subunit alpha, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZR
 
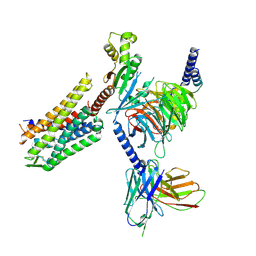 | | GR89,696 bound Kappa Opioid Receptor in complex with gustducin | | Descriptor: | G alpha gustducin protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
7YIT
 
 | |
6CM4
 
 | | Structure of the D2 Dopamine Receptor Bound to the Atypical Antipsychotic Drug Risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, D(2) dopamine receptor, endolysin chimera, ... | | Authors: | Wang, S, Che, T, Levit, A, Shoichet, B.K, Wacker, D, Roth, B.L. | | Deposit date: | 2018-03-02 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone.
Nature, 555, 2018
|
|
7Z2X
 
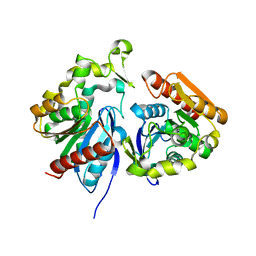 | | Wild-type ferulic acid esterase from Lactobacillus buchneri | | Descriptor: | Ferulic acid esterase | | Authors: | Mogodiniyai, K.K, Reichenbach, T, Kalyani, D.C, Keskitalo, M.M, Divne, C. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the feruloyl esterase from Lentilactobacillus buchneri reveals a novel homodimeric state.
Front Microbiol, 13, 2022
|
|
7Z2U
 
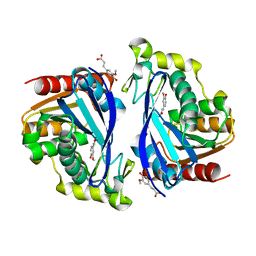 | | Wild-type ferulic acid esterase from Lactobacillus buchneri in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, Ferulic acid esterase | | Authors: | Mogodiniyai, K.K, Reichenbach, T, Kalyani, D.C, Keskitalo, M.M, Divne, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the feruloyl esterase from Lentilactobacillus buchneri reveals a novel homodimeric state.
Front Microbiol, 13, 2022
|
|
7Z2V
 
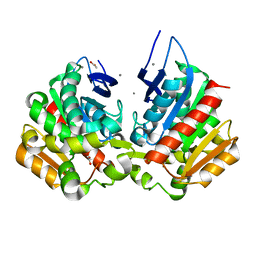 | | Ferulic acid esterase variant S114A from Lactobacillus buchneri | | Descriptor: | ACETATE ION, CALCIUM ION, Ferulic acid esterase | | Authors: | Mogodiniyai, K.K, Reichenbach, T, Kalyani, D.C, Keskitalo, M.M, Divne, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the feruloyl esterase from Lentilactobacillus buchneri reveals a novel homodimeric state.
Front Microbiol, 13, 2022
|
|
5WIV
 
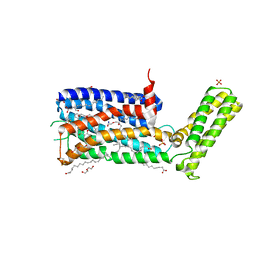 | | Structure of the sodium-bound human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5WIU
 
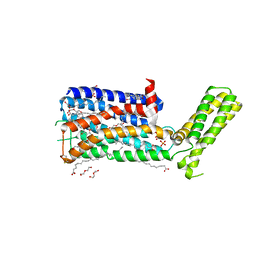 | | Structure of the human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
6LHO
 
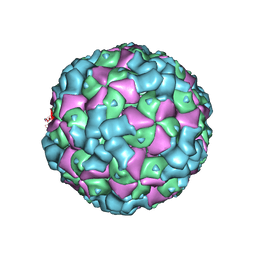 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHK
 
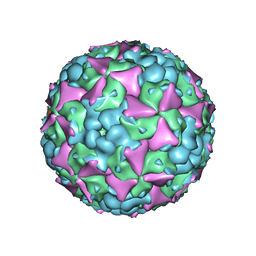 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
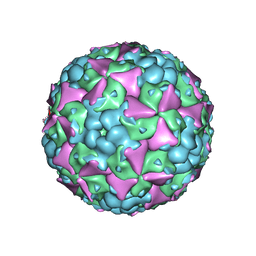 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
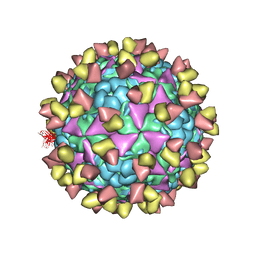 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
