1D1O
 
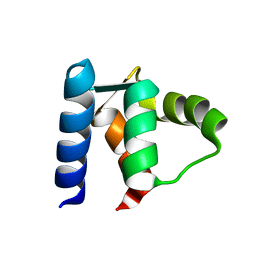 | |
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
7LMW
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 7-methyl-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
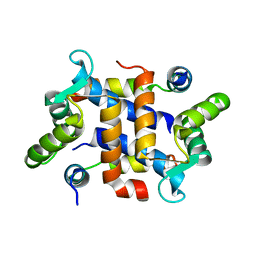
2JTT
 
 | |
1YSM
 
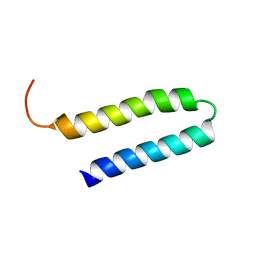 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
2KEB
 
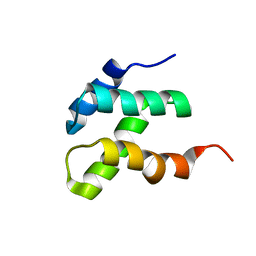 | | NMR solution structure of the N-terminal domain of the DNA polymerase alpha p68 subunit | | Descriptor: | DNA polymerase subunit alpha B | | Authors: | Huang, H, Weiner, B.E, Zhang, H, Fuller, B.E, Gao, Y, Wile, B.M, Chazin, W.J, Fanning, E. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome.
J.Biol.Chem., 285, 2010
|
|
1CLB
 
 | |
4LW1
 
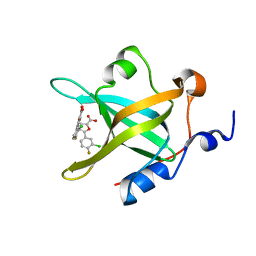 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LWC
 
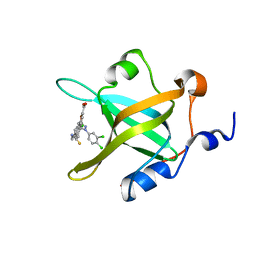 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-[3-chloro-4-({4-[1-(3,4-dichlorophenyl)-1H-pyrazol-5-yl]benzyl}carbamothioyl)phenyl]furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-26 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LUZ
 
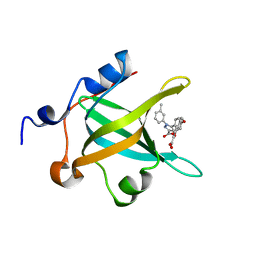 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
5DQO
 
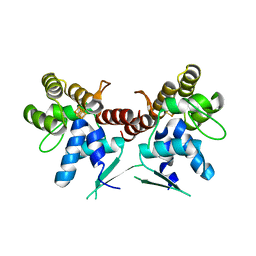 | |
5DBR
 
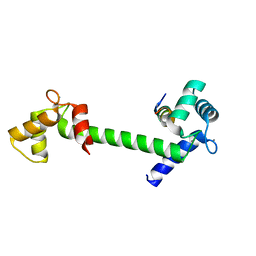 | |
8G9F
 
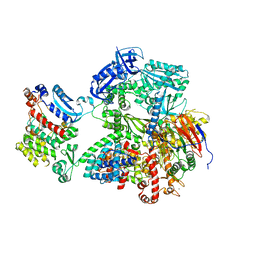 | | Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G9L
 
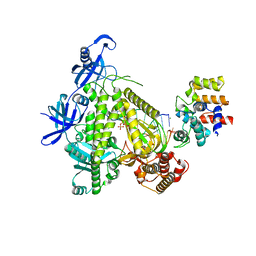 | | DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G9O
 
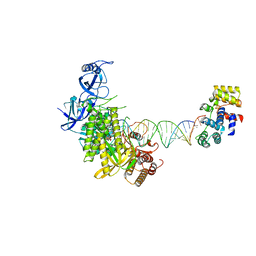 | | Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G9N
 
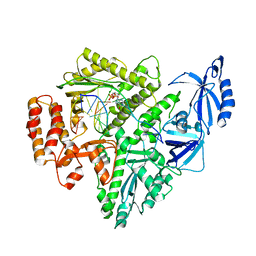 | | Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G99
 
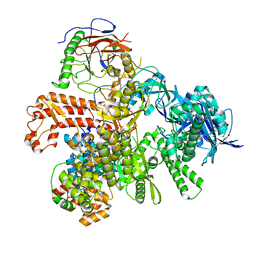 | | Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5E7N
 
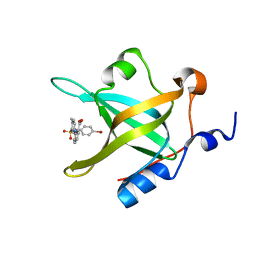 | | Crystal Structure of RPA70N in complex with VU0085636 | | Descriptor: | 2-({3-[(4-bromophenyl)sulfamoyl]-4-methylbenzoyl}amino)benzoic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Gilston, B.A, Patrone, J.D, Pelz, N.F, Bates, B.S, Souza-Fagundes, E.M, Vangamudi, B, Camper, D, Kuznetsov, A, Browning, C.F, Feldkamp, M.D, Olejniczak, E.T, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Identification and Optimization of Anthranilic Acid Based Inhibitors of Replication Protein A.
Chemmedchem, 11, 2016
|
|
1PSB
 
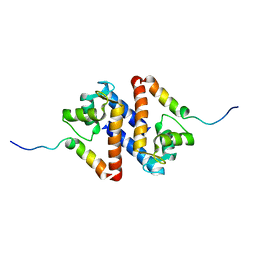 | | Solution structure of calcium loaded S100B complexed to a peptide from N-Terminal regulatory domain of NDR kinase. | | Descriptor: | Ndr Ser/Thr kinase-like protein, S-100 protein, beta chain | | Authors: | Bhattacharya, S, Large, E, Heizmann, C.W, Hemmings, B, Chazin, W.J. | | Deposit date: | 2003-06-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Biochemistry, 42, 2003
|
|
1OQP
 
 | |
2L53
 
 | |
2AMI
 
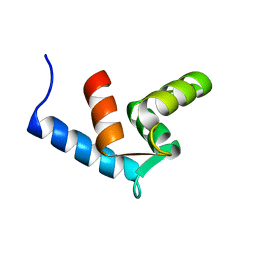 | | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | | Descriptor: | Caltractin | | Authors: | Hu, H.T, Fagan, P.A, Bunick, C.G, Sheehan, J.H, Chazin, W.J. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function.
J.Biol.Chem., 281, 2006
|
|
2KBI
 
 | |
2BAY
 
 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
2KWQ
 
 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
