7QBV
 
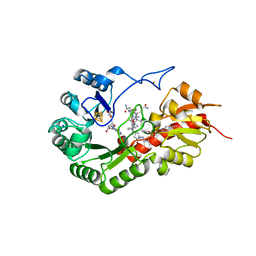 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-adenosyl-L-homocysteine bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
2ZMD
 
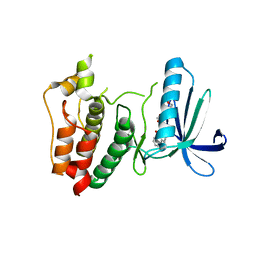 | | Crystal structure of human Mps1 catalytic domain T686A mutant in complex with SP600125 inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Douglas, K.T, Eyers, P.A, Tabernero, L. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125.
J.Biol.Chem., 283, 2008
|
|
3HMN
 
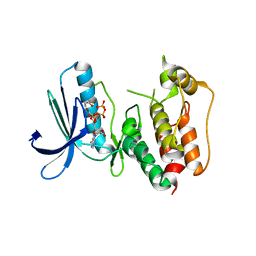 | | Crystal structure of human Mps1 catalytic domain in complex with ATP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
3HMO
 
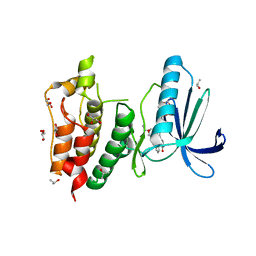 | | Crystal structure of human Mps1 catalytic domain in complex with the inhibitor staurosporine | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK, GLYCEROL, ... | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
3HMP
 
 | | Crystal structure of human Mps1 catalytic domain in complex with a quinazolin ligand Compound 4 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 7-chloro-N-(cyclopropylmethyl)quinazolin-4-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
2ZMC
 
 | | Crystal structure of human mitotic checkpoint kinase Mps1 catalytic domain apo form | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Douglas, K.T, Eyers, P.A, Tabernero, L. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125.
J.Biol.Chem., 283, 2008
|
|
8AI1
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI4
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI3
 
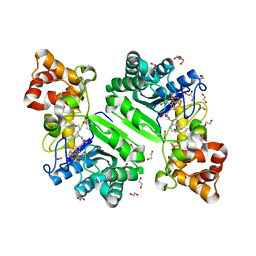 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-methionine bound | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Chavas, L.M.G, Legrand, P, Polsinelli, I, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI2
 
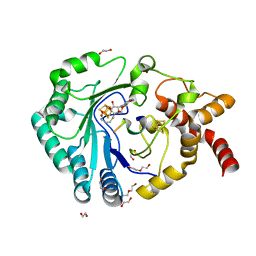 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI5
 
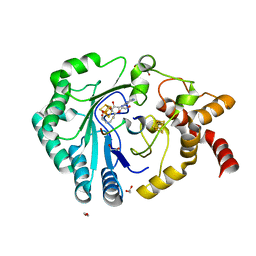 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
4NC5
 
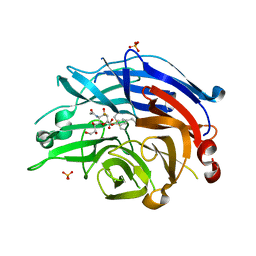 | | Human sialidase 2 in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, PHOSPHATE ION, Sialidase-2 | | Authors: | Buchini, S, Gallat, F.-X, Greig, I.R, Kim, J.-H, Wakatsuki, S, Chavas, L.M.G, Withers, S.G. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.513 Å) | | Cite: | Tuning mechanism-based inactivators of neuraminidases: mechanistic and structural insights.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NYR
 
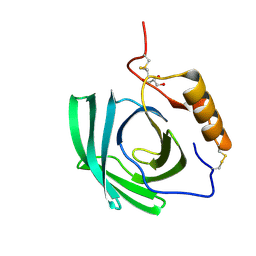 | | In-vivo crystallisation (midguts of a viviparous cockroach) and structure at 2.5 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Coussens, N.P, Gallat, F.-X, Ramaswamy, S, Yagi, K, Tobe, S.S, Stay, B, Chavas, L.M.G. | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
4NCS
 
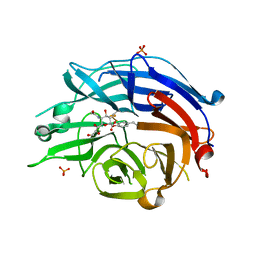 | | Human sialidase 2 in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-2,3-bis(fluoranyl)-4-oxidanyl-6-[(1S,2S)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, PHOSPHATE ION, Sialidase-2 | | Authors: | Buchini, S, Gallat, F.-X, Greig, I.R, Kim, J.-H, Wakatsuki, S, Chavas, L.M.G, Withers, S.G. | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Tuning mechanism-based inactivators of neuraminidases: mechanistic and structural insights.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NYQ
 
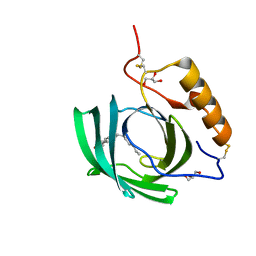 | | In-vivo crystallisation (midguts of a viviparous cockroach) and structure at 1.2 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Coussens, N.P, Gallat, F.-X, Ramaswamy, S, Yagi, K, Tobe, S.S, Stay, B, Chavas, L.M.G. | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
