1XG7
 
 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
1ZGH
 
 | | Methionyl-tRNA formyltransferase from Clostridium thermocellum | | Descriptor: | Methionyl-tRNA formyltransferase, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Kataeva, I, Xu, H, Zhao, M, Chang, J, Liu, Z, Chen, L, Tempel, W, Habel, J, Zhou, W, Lee, D, Lin, D, Chang, S, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionyl-tRNA formyltransferase from Clostridium thermocellum
To be published
|
|
3K6G
 
 | | Crystal structure of Rap1 and TRF2 complex | | Descriptor: | Telomeric repeat-binding factor 2, Telomeric repeat-binding factor 2-interacting protein 1 | | Authors: | Chen, Y, Rai, R, Yang, Y.T, Zheng, H, Chang, S, Lei, M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8JYU
 
 | | Acyl-ACP Synthetase structure bound to Decanoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, DECANOIC ACID, ... | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Acyl-ACP Synthetase structure bound to Decanoyl-AMP
To Be Published
|
|
6U07
 
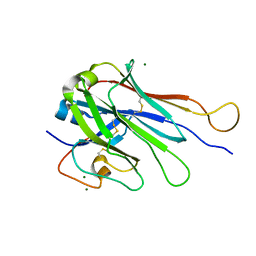 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
3GMF
 
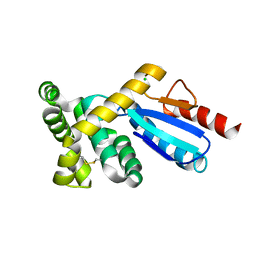 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3IA1
 
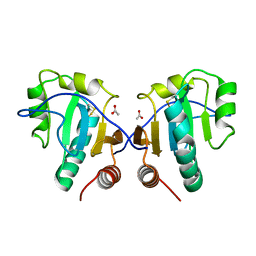 | | Crystal structure of thio-disulfide isomerase from Thermus thermophilus | | Descriptor: | ACETATE ION, Thio-disulfide isomerase/thioredoxin | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of thio-disulfide isomerase from Thermus thermophilus
To be Published
|
|
3IJ6
 
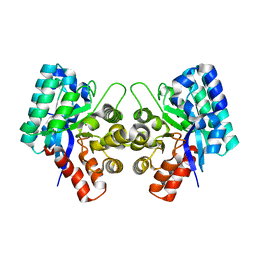 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | Descriptor: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
3I83
 
 | | Crystal structure of 2-dehydropantoate 2-reductase from Methylococcus capsulatus | | Descriptor: | 2-dehydropantoate 2-reductase, ACETIC ACID | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Sampathkumar, P, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase from Methylococcus capsulatus
To be Published
|
|
3IMH
 
 | | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus NCFM | | Descriptor: | CHLORIDE ION, GLYCEROL, Galactose-1-epimerase, ... | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus
To be Published
|
|
3IPL
 
 | | CRYSTAL STRUCTURE OF o-succinylbenzoic acid-CoA ligase FROM Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 2-succinylbenzoate--CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of o-succinylbenzoic acid-CoA ligase from Staphylococcus aureus
To be Published
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
3HCW
 
 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | Descriptor: | GLYCEROL, Maltose operon transcriptional repressor | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
3GRZ
 
 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3H7L
 
 | | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus | | Descriptor: | ENDOGLUCANASE, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-27 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus
To be Published
|
|
3HN2
 
 | | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens GS-15 | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens
To be Published
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
3GG2
 
 | | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate | | Descriptor: | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
To be Published
|
|
3GKB
 
 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3E5R
 
 | |
3I9F
 
 | | Crystal structure of a putative type 11 methyltransferase from Sulfolobus solfataricus | | Descriptor: | Putative type 11 methyltransferase, ZINC ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative type 11 methyltransferase from Sulfolobus solfataricus
To be Published
|
|
3O8L
 
 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
