6JAR
 
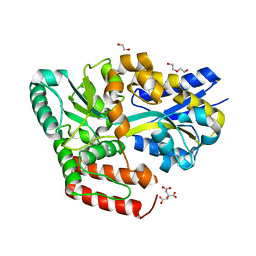 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R49A in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6JAH
 
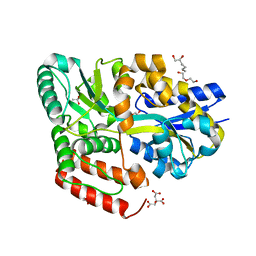 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with glucose | | Descriptor: | (2S)-heptane-1,2,7-triol, 1,2-ETHANEDIOL, ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
2AVK
 
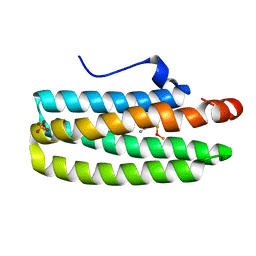 | | met-azido-DcrH-Hr | | Descriptor: | MONOAZIDO-MU-OXO-DIIRON, SULFATE ION, hemerythrin-like domain protein DcrH | | Authors: | Isaza, C.E, Silaghi-Dumitrescu, R, Iyer, R.B, Kurtz, D.M, Chan, M.K. | | Deposit date: | 2005-08-30 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Basis for O(2) Sensing by the Hemerythrin-like Domain of a Bacterial Chemotaxis Protein: Substrate Tunnel and Fluxional N Terminus.
Biochemistry, 45, 2006
|
|
2AWC
 
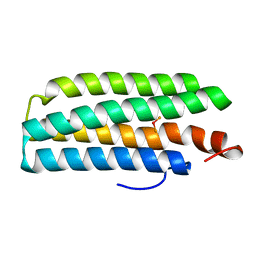 | | deoxy-DcrH-Hr | | Descriptor: | MU-OXO-DIIRON, hemerythrin-like domain protein DcrH | | Authors: | Isaza, C.E, Silaghi-Dumitrescu, R, Iyer, R.B, Kurtz, D.M, Chan, M.K. | | Deposit date: | 2005-08-31 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for O(2) Sensing by the Hemerythrin-like Domain of a Bacterial Chemotaxis Protein: Substrate Tunnel and Fluxional N Terminus
Biochemistry, 45, 2006
|
|
2AWY
 
 | | met-DcrH-Hr | | Descriptor: | CALCIUM ION, CHLORIDE ION, MU-OXO-DIIRON, ... | | Authors: | Isaza, C.E, Silaghi-Dumitrescu, R, Iyer, R.B, Kurtz, D.M, Chan, M.K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for O(2) Sensing by the Hemerythrin-like Domain of a Bacterial Chemotaxis Protein: Substrate Tunnel and Fluxional N Terminus
Biochemistry, 45, 2006
|
|
3CF4
 
 | | Structure of the CODH component of the M. barkeri ACDS complex | | Descriptor: | ACETIC ACID, Acetyl-CoA decarboxylase/synthase alpha subunit, Acetyl-CoA decarboxylase/synthase epsilon subunit, ... | | Authors: | Gong, W, Hao, B, Wei, Z, Ferguson Jr, D.J, Tallant, T, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2008-03-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1TV4
 
 | | Crystal structure of the sulfite MtmB complex | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1, SULFATE ION | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
7T3F
 
 | | Development of BRD4 inhibitors as arsenicals antidotes | | Descriptor: | 4-fluoro-3-methyl-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Wu, M, Yatchang, M, Mathew, B, Zhai, L, Ruiz, P, Bostwick, R, Augelli-Szafran, C.E, Suto, M.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Development of BRD4 inhibitors as anti-inflammatory agents and antidotes for arsenicals.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
1XEM
 
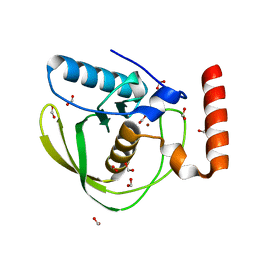 | | High Resolution Crystal Structure of Escherichia coli Zinc- Peptide Deformylase bound to formate | | Descriptor: | FORMIC ACID, Peptide deformylase, ZINC ION | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1TV3
 
 | | Crystal structure of the N-methyl-hydroxylamine MtmB complex | | Descriptor: | 5-(HYDROXY-METHYL-AMINO)-3-METHYL-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
1TV2
 
 | | Crystal structure of the hydroxylamine MtmB complex | | Descriptor: | 5-HYDROXYAMINO-3-METHYL-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
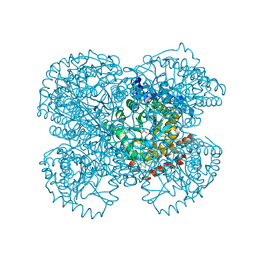
1HUZ
 
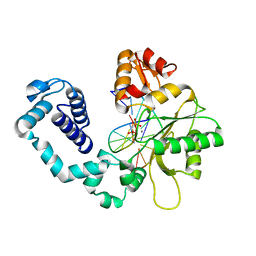 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
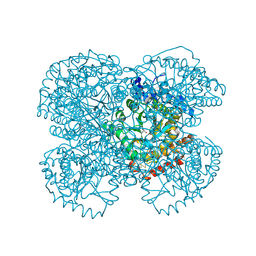
1HUO
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1XEO
 
 | | High Resolution Crystals Structure of Cobalt- Peptide Deformylase Bound To Formate | | Descriptor: | COBALT (II) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEN
 
 | | High Resolution Crystal Structure of Escherichia coli Iron- Peptide Deformylase Bound To Formate | | Descriptor: | FE (III) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
2LRN
 
 | | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp. | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-10 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp.
To be Published
|
|
2LST
 
 | | Solution structure of a thioredoxin from Thermus thermophilus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thioredoxin from Thermus thermophilus
To be Published
|
|
2LRT
 
 | | Solution structure of the uncharacterized thioredoxin-like protein BVU_1432 from Bacteroides vulgatus | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the uncharacterized thioredoxin-like protein BVU_1432 from Bacteroides vulgatus
To be Published
|
|
2LS8
 
 | | Solution structure of human C-type lectin domain family 4 member D | | Descriptor: | C-type lectin domain family 4 member D | | Authors: | Harris, R, Gaudette, J, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human C-type lectin domain family 4 member D
To be Published
|
|
2LS5
 
 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
4PFQ
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 029763. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 0299763.
to be published
|
|
4NS1
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
To be Published
|
|
4NSN
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry | | Descriptor: | ADENINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry.
To be Published
|
|
4F3X
 
 | | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aldehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD
To be Published
|
|
4FFU
 
 | | CRYSTAL STRUCTURE OF putative MaoC-like (monoamine oxidase-like) protein, similar to NodN from Sinorhizo Bium meliloti 1021 | | Descriptor: | CHLORIDE ION, oxidase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC-like (monoamine oxidase-like) protein, similar to NodN from Sinorhizo
Bium meliloti 1021
To be Published
|
|
