8BNZ
 
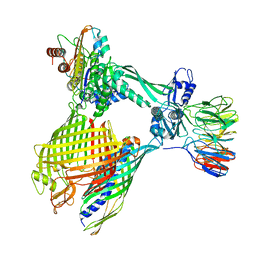 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
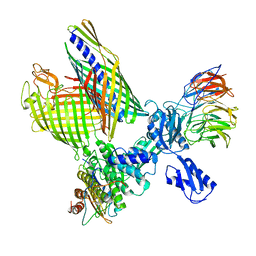 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BQF
 
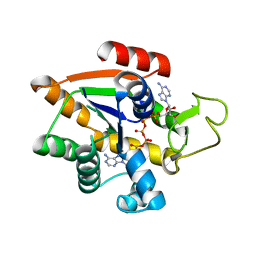 | | Adenylate Kinase L107I MUTANT | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Scheerer, D, Adkar, B.V, Bhattacharyya, S, Levy, D, Iljina, M, Iljina, I, Dym, O, Haran, G, Shakhnovich, E.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4KAL
 
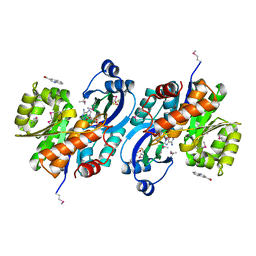 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinoline-3-carboxylic acid | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinoline-3-carboxylic acid
To be Published
|
|
8BLX
 
 | | Engineered Fructosyl Peptide Oxidase - X02A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X02A), ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BJY
 
 | | Engineered Fructosyl Peptide Oxidase - X02B mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X02B), GLYCEROL | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8BLZ
 
 | | Engineered Fructosyl Peptide Oxidase - D02 mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (D02), GLYCEROL, ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
8P1U
 
 | | Structure of divisome complex FtsWIQLB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Yang, L, Chang, S, Tang, D, Dong, H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
8BMU
 
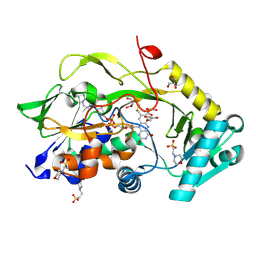 | | Engineered Fructosyl Peptide Oxidase - X04 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase mutant (X04), ... | | Authors: | Estiri, H, Bhattacharya, S, Rodriguez-Buitrago, J.A, Parisini, E. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tailoring FPOX enzymes for enhanced stability and expanded substrate recognition.
Sci Rep, 13, 2023
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8CGX
 
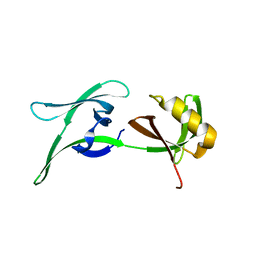 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using XDS | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CGY
 
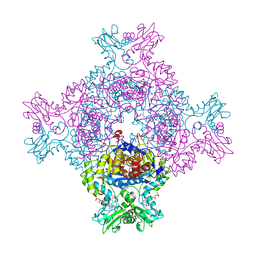 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; XDS processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
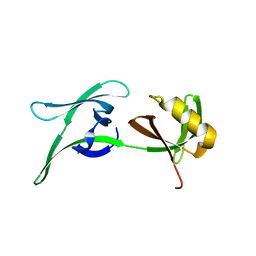 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
5KN9
 
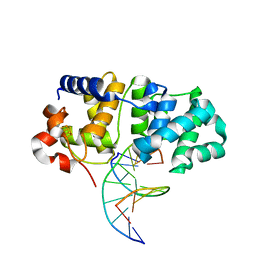 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
4ZWN
 
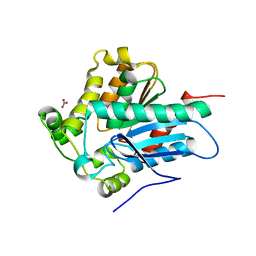 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
5KN8
 
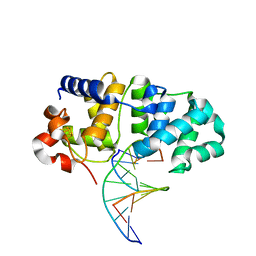 | | MutY N-terminal domain in complex with undamaged DNA | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
6GWU
 
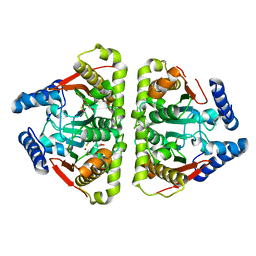 | | Carbonic anhydrase CaNce103p from Candida albicans | | Descriptor: | BETA-MERCAPTOETHANOL, Carbonic anhydrase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brynda, J, Dostal, J, Heidingsfeld, O, Machacek, S, Blaha, J, Pichova, I. | | Deposit date: | 2018-06-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of carbonic anhydrase CaNce103p from the pathogenic yeast Candida albicans.
BMC Struct. Biol., 18, 2018
|
|
8C53
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; CrystFEL processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C5K
 
 | | HEX-1 (in cellulo, in situ) crystallized and diffracted in High Five cells. Growth and SX data collection at 296 K on CrystalDirect plates | | Descriptor: | Woronin body major protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C51
 
 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD4
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo (cytosol), diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD6
 
 | | structure of HEX-1 (cyto V2) from N. crassa grown in living insect cells, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
5DK6
 
 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
8JYU
 
 | | Acyl-ACP Synthetase structure bound to Decanoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, DECANOIC ACID, ... | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Acyl-ACP Synthetase structure bound to Decanoyl-AMP
To Be Published
|
|
