6CHK
 
 | | Crystal structure of LacI family transcriptional regulator from Lactobacillus casei, Target EFI-512911, with bound TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Shabalin, I.G, Kowiel, M, Porebski, P.J, Minor, W, Jaskolski, M, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative, E.F.I. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Automatic recognition of ligands in electron density by machine learning.
Bioinformatics, 35, 2019
|
|
7E4J
 
 | | X-ray crystal structure of VapB12 antitoxin from mycobacterium tuberculosis in space group P41. | | Descriptor: | Antitoxin, ZINC ION | | Authors: | Pratap, S, Megta, A.K, Talwar, S, Chandresh, S, Pandey, A.K, Krishnan, V. | | Deposit date: | 2021-02-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray crystal structure of VapB12 antitoxin from mycobacterium tuberculosis in space group P41.
To be published
|
|
4ZJM
 
 | | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein LpqH, ... | | Authors: | Arbing, M.A, Chan, S, Kuo, E, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763)
To Be Published
|
|
5DMH
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
4DPH
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P65 and NADPH | | Descriptor: | 2,4-diamino-6-methyl-5-[3-(2,4,5-trichlorophenoxy)propyloxy]pyrimidine, BETA-MERCAPTOETHANOL, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5DM3
 
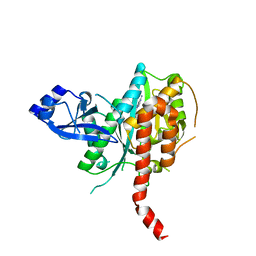 | | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-glutamine synthetase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP
To be published
|
|
3CRN
 
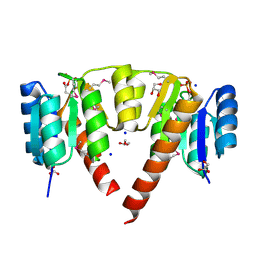 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, Response regulator receiver domain protein, CheY-like, ... | | Authors: | Hiner, R.L, Toro, R, Patskovsky, Y, Freeman, J, Chang, S, Smith, D, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1.
To be Published
|
|
5LP5
 
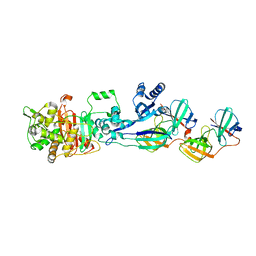 | | Complex between Penicillin-Binding Protein (PBP2) and MreC from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), Rod shape-determining protein (MreC) | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Hicham, S, Hardouin, P, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
4YEC
 
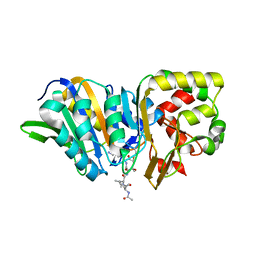 | |
3CZ8
 
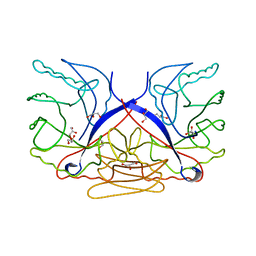 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
4Y9T
 
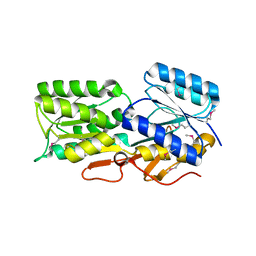 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GLUCOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, ABC transporter, solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7JSN
 
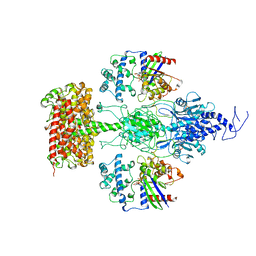 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | Deposit date: | 2020-08-15 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
8A3L
 
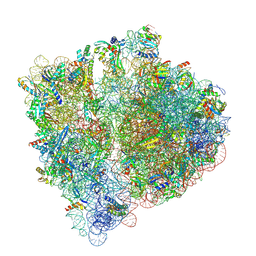 | | Structural insights into the binding of bS1 to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | D'Urso, G, Chat, S, Gillet, R, Giudice, E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the binding of bS1 to the ribosome.
Nucleic Acids Res., 51, 2023
|
|
5XO3
 
 | |
5ABH
 
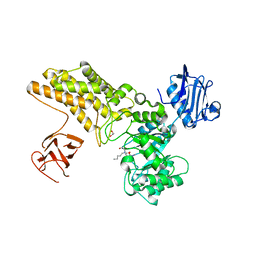 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5CYU
 
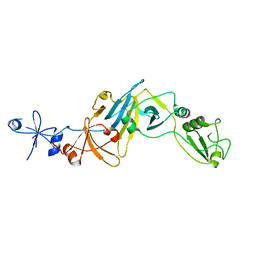 | | Structure of the soluble domain of EccB1 from the Mycobacterium smegmatis ESX-1 secretion system. | | Descriptor: | Conserved membrane protein | | Authors: | Arbing, M.A, Chan, S, Kahng, S, Kim, J, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system.
Bmc Struct.Biol., 16, 2016
|
|
3CG0
 
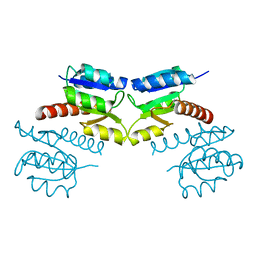 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
5EK9
 
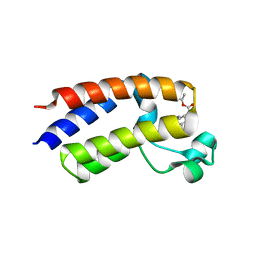 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline inhibitor | | Descriptor: | Bromodomain-containing protein 2, propan-2-yl ~{N}-[(2~{S},4~{R})-1-ethanoyl-6-(furan-2-yl)-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Picaud, S, Fedorov, O, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
6D7R
 
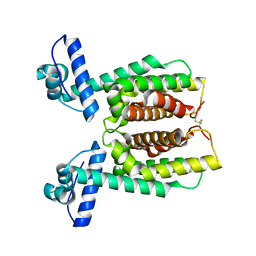 | | Crystal structure of an inactivate variant of the quorum-sensing master regulator HapR from protease-deficient non-O1, non-O139 Vibrio cholerae strain V2 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Cruite, J.T, Succo, P, Raychaudhuri, S, Kull, F.J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an inactive variant of the quorum-sensing master regulator HapR from the protease-deficient non-O1, non-O139 Vibrio cholerae strain V2.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5DTE
 
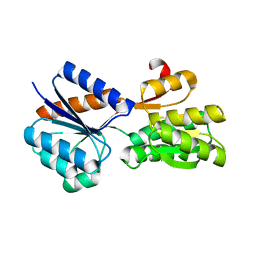 | | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose | | Descriptor: | Monosaccharide-transporting ATPase, beta-D-allopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an ABC transporter periplasmic solute binding protein (IPR025997) from Actinobacillus succinogenes 130z(Asuc_0081, TARGET EFI-511065) with bound D-allose
To be published
|
|
5ABE
 
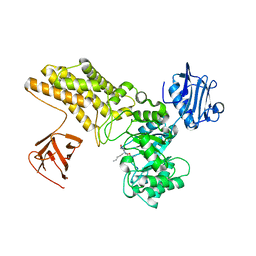 | | Structure of GH84 with ligand | | Descriptor: | 2-[(2S,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5ABF
 
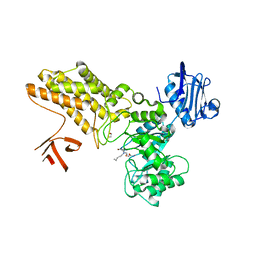 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S,3R,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DKV
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose | | Descriptor: | ABC transporter substrate binding protein (Ribose), alpha-D-tagatopyranose | | Authors: | Yadava, U, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Agrobacterium vitis(Avis_5339, TARGET EFI-511225) bound with alpha-D-Tagatopyranose
To be published
|
|
6DXP
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
5BR1
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
