5VOD
 
 | |
7DCK
 
 | | Crystal structure of phosphodiesterase tw9814 | | Descriptor: | Lactamase_B domain-containing protein, MANGANESE (II) ION | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Park, S.B, Cha, S.S, Lee, W. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and functional identification of the uncharacterized metallo-beta-lactamase superfamily protein TW9814 as a phosphodiesterase with unique metal coordination.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5VOB
 
 | |
6DG8
 
 | | Full-length 5-HT3A receptor in a serotonin-bound conformation- State 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, SEROTONIN, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3Areceptor.
Nature, 563, 2018
|
|
6DG7
 
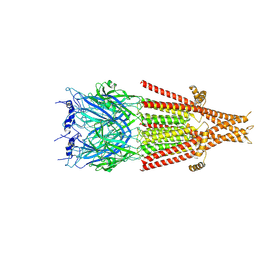 | | Full-length 5-HT3A receptor in a serotonin-bound conformation- State 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, SEROTONIN, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3Areceptor.
Nature, 563, 2018
|
|
2LMA
 
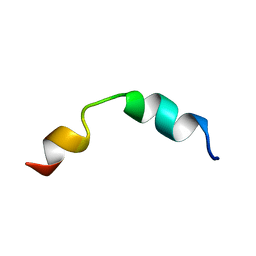 | | Solution structure of CD4+ T cell derived peptide Thp5 | | Descriptor: | Thp5 peptide | | Authors: | Pandey, N.K, Khan, M.M, Chatterjee, S, Dwivedi, V.P, Tousif, S, Das, J, Van Kaer, L. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CD4+ T cell derived novel peptide Thp5 induces IL-4 production in CD4+ T cells to direct T helper 2 cell differentiation
J.Biol.Chem., 2011
|
|
5X9V
 
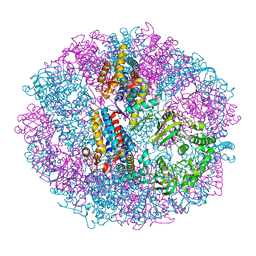 | | Crystal structure of group III chaperonin in the Closed state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome, ... | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
4J4K
 
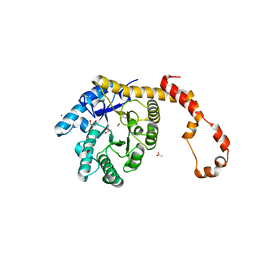 | | Crystal structure of glucose isomerase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Xylose isomerase, ... | | Authors: | Kim, M.K, An, Y.J, Lee, S, Jeong, C.S, Cha, S.S. | | Deposit date: | 2013-02-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glucose isomerase
To be Published
|
|
5Z8A
 
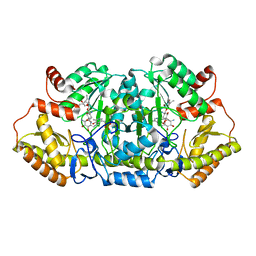 | | Crystal structure of GenB1 from Micromonospora echinospora in complex with JI-20A and PLP (external aldimine) | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-2-[(1~{S},2~{S},3~{R},4~{S},6~{R})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-6-(aminomethyl)-3-azany l-4,5-bis(oxidanyl)oxan-2-yl]oxy-4,6-bis(azanyl)-2-oxidanyl-cyclohexyl]oxy-5-methyl-4-(methylamino)oxane-3,5-diol, C-6' aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hong, S.K, Cha, S.S. | | Deposit date: | 2018-01-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
7E3Z
 
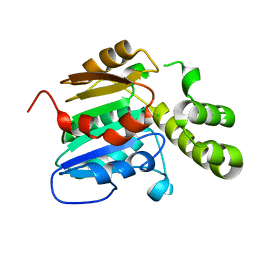 | | Non-Ribosomal Peptide Synthetases, Thioesterase | | Descriptor: | CHLORIDE ION, thioesterase | | Authors: | Jung, Y.E, Cha, S.S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unprecedented Noncanonical Features of the Nonlinear Nonribosomal Peptide Synthetase Assembly Line for WS9326A Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5Z83
 
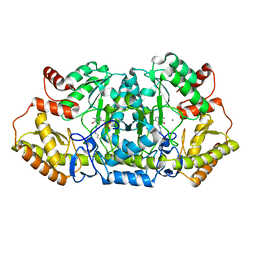 | |
7YGK
 
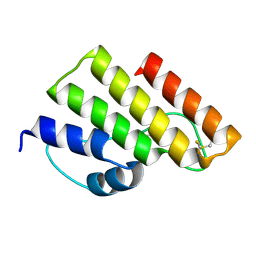 | |
3KMM
 
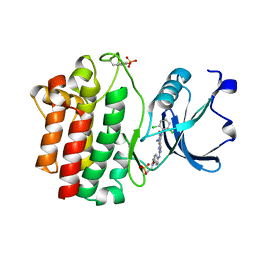 | | Structure of human LCK kinase with a small molecule inhibitor | | Descriptor: | 3-(2,6-dichlorophenyl)-7-({4-[2-(diethylamino)ethoxy]phenyl}amino)-1-methyl-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Graves, B.J, Surgenor, A, Harris, W, Smith, I, Orchard, S, Flotow, H, Murray, E. | | Deposit date: | 2009-11-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human LCK kinase with a small molecule inhibitor
To be Published
|
|
4TYM
 
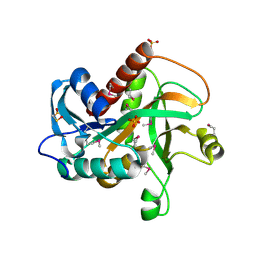 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
3V1U
 
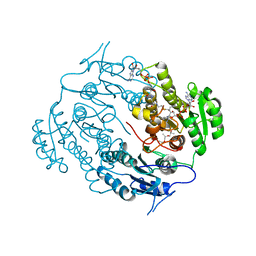 | | Crystal structure of a beta-ketoacyl reductase FabG4 from Mycobacterium tuberculosis H37Rv complexed with NAD+ and Hexanoyl-CoA at 2.5 Angstrom resolution | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, HEXANOYL-COENZYME A, ... | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
3LQ7
 
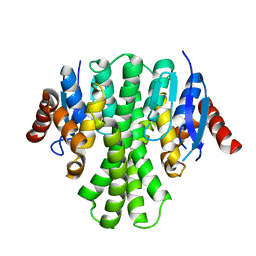 | | Crystal structure of glutathione s-transferase from agrobacterium tumefaciens str. c58 | | Descriptor: | Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Agrobacterium Tumefaciens
To be Published
|
|
5F1G
 
 | | Crystal structure of AmpC BER adenylylated in the cytoplasm | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Beta-lactamase, ... | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5F1F
 
 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5GGW
 
 | | Crystal structure of Class C beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5GMX
 
 | | Crystal structure of a family VIII carboxylesterase | | Descriptor: | Carboxylesterase, phenylmethanesulfonic acid | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5H4U
 
 | | Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | An, Y.J, Hong, S.K, Cha, S.S. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and Structural Characterization of a Thermo-Tolerant, Cold-Active, and Acidic Endo-beta-1,4-glucanase from Antarctic Springtail, Cryptopygus antarcticus.
J. Agric. Food Chem., 65, 2017
|
|
4YEU
 
 | |
7R1M
 
 | | Crystal structure of ExsFA, a Bacillus cereus spore exosporium protein | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Exosporium protein, ... | | Authors: | Brear, P, Schack, S, Christie, G. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of ExsFA, a Bacillus cereus spore exosporium protein"
To Be Published
|
|
7R5I
 
 | | High resolution Crystal structure of ExsFA, a Bacillus cereus spore exosporium protein | | Descriptor: | CALCIUM ION, Exosporium protein, GLN-GLU-ASP-PHE-SER-SER-ASP-SER-SER-PHE-SER, ... | | Authors: | Brear, P, Schack, S, Christie, G. | | Deposit date: | 2022-02-10 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of ExsFA, a Bacillus cereus spore exosporium protein"
To Be Published
|
|
3L18
 
 | |
