2RCY
 
 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
2EXG
 
 | | Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain | | Descriptor: | (5R)-2-SULFANYL-5-[4-(TRIFLUOROMETHYL)BENZYL]-1,3-THIAZOL-4-ONE, Afadin | | Authors: | Joshi, M, Vargas, C, Boisguerin, P, Krause, G, Schade, M, Oschkinat, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
3SHM
 
 | |
4BDI
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-acetyl-N-(5-methylpyridin-2-yl)piperidine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDC
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(furan-2-ylmethyl)quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
2R77
 
 | | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum | | Descriptor: | Phosphatidylethanolamine-binding protein, putative | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Crombette, L, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum.
To be Published
|
|
3DM1
 
 | | Crystal structure of the complex of human chromobox homolog 3 (CBX3) with peptide | | Descriptor: | Chromobox protein homolog 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
4BDD
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
3C5K
 
 | | Crystal structure of human HDAC6 zinc finger domain | | Descriptor: | Histone deacetylase 6, ZINC ION | | Authors: | Dong, A, Ravichandran, M, Schuetz, A, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human HDAC6 zinc finger domain.
To be Published
|
|
3Q5L
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-{[2-(pyrrolidin-1-yl)ethyl]amino}-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
To be Published
|
|
3Q5K
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-{[2-(methylsulfanyl)ethyl]amino}benzamide, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor
To be Published
|
|
4A94
 
 | | Structure of the carboxypeptidase inhibitor from Nerita versicolor in complex with human CPA4 | | Descriptor: | CARBOXYPEPTIDASE A4, CARBOXYPEPTIDASE INHIBITOR, NITRATE ION, ... | | Authors: | Covaleda, G, Alonso, M, Chavez, M.A, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-28 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Novel Metallo-Carboxypeptidase Inhibitor from the Marine Mollusk Nerita Versicolor in Complex with Human Carboxypeptidase A4.
J.Biol.Chem., 287, 2012
|
|
3C5I
 
 | | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520 | | Descriptor: | CALCIUM ION, CHOLINE ION, Choline kinase, ... | | Authors: | Wernimont, A.K, Hills, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520.
To be Published
|
|
1LB7
 
 | | IGF-F1-1, A PEPTIDE ANTAGONIST OF IGF-1 | | Descriptor: | IGF-1 ANTAGONIST F1-1 | | Authors: | Deshayes, K, Schaffer, M.L, Skelton, N.J, Nakamura, G.R, Kadkhodayan, S, Sidhu, S.S. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Rapid identification of small binding motifs with high-throughput phage display: discovery of peptidic antagonists of IGF-1 function.
Chem.Biol., 9, 2002
|
|
1XEO
 
 | | High Resolution Crystals Structure of Cobalt- Peptide Deformylase Bound To Formate | | Descriptor: | COBALT (II) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
3SMQ
 
 | | Crystal structure of protein arginine methyltransferase 3 | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | Authors: | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
3G2J
 
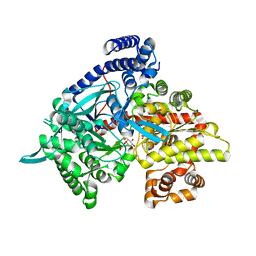 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-(hydroxyacetyl)-beta-D-glucopyranosylamine | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
1XEM
 
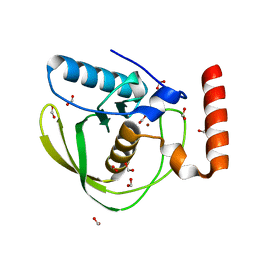 | | High Resolution Crystal Structure of Escherichia coli Zinc- Peptide Deformylase bound to formate | | Descriptor: | FORMIC ACID, Peptide deformylase, ZINC ION | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
3Q5J
 
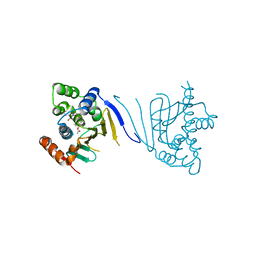 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
3V0R
 
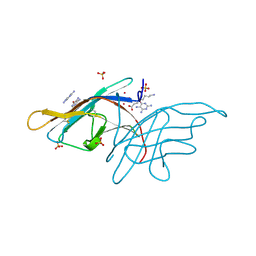 | | Crystal structure of Alternaria alternata allergen Alt a 1 | | Descriptor: | 2,5,6-triaminopyrimidin-4-ol, 8-aminooctanoic acid, Major allergen Alt a 1, ... | | Authors: | Chruszcz, M, Solberg, R, Osinski, T, Chapman, M.D, Minor, W. | | Deposit date: | 2011-12-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternaria alternata allergen Alt a 1: a unique beta-barrel protein dimer found exclusively in fungi.
J.Allergy Clin.Immunol., 130, 2012
|
|
3PHD
 
 | | Crystal structure of human HDAC6 in complex with ubiquitin | | Descriptor: | Histone deacetylase 6, Polyubiquitin, ZINC ION | | Authors: | Dong, A, Qui, W, Ravichandran, M, Schuetz, A, Loppnau, P, Li, F, Mackenzie, F, Kozieradzki, I, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Aggregates Are Recruited to Aggresome by Histone Deacetylase 6 via Unanchored Ubiquitin C Termini.
J.Biol.Chem., 287, 2012
|
|
3CXG
 
 | | Crystal structure of Plasmodium falciparum thioredoxin, PFI0790w | | Descriptor: | GLYCEROL, Putative thioredoxin, SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Hills, T, Pizarro, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-24 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Plasmodium falciparum thioredoxin, PFI0790w.
To be Published
|
|
3PKJ
 
 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
1LT0
 
 | | Crystal structure of the CN-bound BjFixL heme domain | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-20 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
