4Z0F
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038(6CW)] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0D
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038Trp] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
2LQY
 
 | | Structure and orientation of the gH625-644 membrane interacting region of herpes simplex virus type 1 in a membrane mimetic system. | | Descriptor: | Envelope glycoprotein H | | Authors: | Isernia, C, Galdiero, S, Russo, L, Falanga, A, Cantisani, M, Vitiello, M, Fattorusso, R, Malgieri, G, Galdiero, M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Orientation of the gH625-644 Membrane Interacting Region of Herpes Simplex Virus Type 1 in a Membrane Mimetic System.
Biochemistry, 51, 2012
|
|
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
1J5H
 
 | | Solution Structure of Apo-Neocarzinostatin | | Descriptor: | Apo-Neocarzinostatin | | Authors: | Urbaniak, M.D, Muskett, F.W, Finucane, M.D, Caddick, S, Woolfson, D.N. | | Deposit date: | 2002-05-02 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore
Biochemistry, 41, 2002
|
|
1J5I
 
 | | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore | | Descriptor: | 2-HYDROXY-7-METHOXY-5-METHYL-NAPHTHALENE-1-CARBOXYLIC ACID MESO-2,5-DIHYDROXY-CYCLOPENT-3-ENYL ESTER, PROTEIN (Apo-Neocarzinostatin) | | Authors: | Urbaniak, M.D, Muskett, F.W, Finucane, M.D, Caddick, S, Woolfson, D.N. | | Deposit date: | 2002-05-02 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Novel Chromoprotein Derived from Apo-Neocarzinostatin and a Synthetic Chromophore
Biochemistry, 41, 2002
|
|
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
2MBT
 
 | | NMR study of PaDsbA | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Rimmer, K, Mohanty, B, Scanlon, M.J. | | Deposit date: | 2013-08-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
2LZ5
 
 | | Solution structure of a Novel Alpha-Conotoxin TxIB | | Descriptor: | Conotoxin_TxIB | | Authors: | Luo, S, Zhangsun, D, Wu, Y, Zhu, X, Hu, Y, McIntyre, M, Christensen, S, Akcan, M, Craik, D, McIntosh, J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a novel alpha-conotoxin from conus textile that selectively targets alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors.
J.Biol.Chem., 288, 2013
|
|
2NDO
 
 | | Structure of EcDsbA-sulfonamide1 complex | | Descriptor: | 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Williams, M.L, Doak, B.C, Vazirani, M, Ilyichova, O, Wang, G, Bermel, W, Simpson, J.S, Chalmers, D.K, King, G.F, Mobli, M, Scanlon, M.J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
2MPM
 
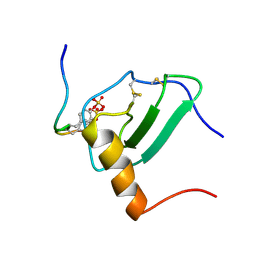 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
2BAG
 
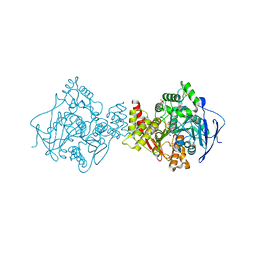 | | 3D Structure of Torpedo californica acetylcholinesterase complexed with Ganstigmine | | Descriptor: | 1S,3AS,8AS-TRIMETHYL-1-OXIDO-1,2,3,3A,8,8A-HEXAHYDROPYRROLO[2,3-B]INDOL-5-YL 2-ETHYLPHENYLCARBAMATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lamba, D, Bartolucci, C, Siotto, M, Racchi, M, Villetti, G, Delcanale, M. | | Deposit date: | 2005-10-14 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Determinants of Torpedo californica Acetylcholinesterase Inhibition by the Novel and Orally Active Carbamate Based Anti-Alzheimer Drug Ganstigmine (CHF-2819)
J.Med.Chem., 49, 2006
|
|
7L7C
 
 | |
7LHP
 
 | |
7L76
 
 | |
4EZA
 
 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
4I01
 
 | | Structure of the mutant Catabolite gen activator protein V140L | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4I0A
 
 | | structure of the mutant Catabolite gene activator protein V132A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, GLYCEROL | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4I09
 
 | | structure of the mutant Catabolite gene activator protein V132L | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4HZF
 
 | | structure of the wild type Catabolite gene Activator Protein | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, GLYCEROL, ... | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4I02
 
 | | structure of the mutant Catabolite gene activator protein V140A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4I0B
 
 | | structure of the mutant Catabolite gene activator protein H160L | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
2KKK
 
 | |
3OVN
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 1-methyl-3-(thiophen-2-yl)-1H-pyrazol-5-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Deadman, J.J, Rhodes, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
2MO5
 
 | | hIFABP-oleate complex | | Descriptor: | Fatty acid-binding protein, intestinal, OLEIC ACID | | Authors: | Patil, R, Mohanty, B, Headey, S, Porter, C, Scanlon, M. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of oleate bound human intestinal fatty acid binding protein
To be Published
|
|
