3BEW
 
 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
3SWF
 
 | | CNGA1 621-690 containing CLZ domain | | Descriptor: | ZINC ION, cGMP-gated cation channel alpha-1 | | Authors: | Shuart, N.G, Haitin, Y, Camp, S.S, Black, K.D, Zagotta, W.N. | | Deposit date: | 2011-07-13 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels.
Nat Commun, 2, 2011
|
|
3SWY
 
 | | CNGA3 626-672 containing CLZ domain | | Descriptor: | Cyclic nucleotide-gated cation channel alpha-3 | | Authors: | Shuart, N.G, Haitin, Y, Camp, S.S, Black, K.D, Zagotta, W.N. | | Deposit date: | 2011-07-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels.
Nat Commun, 2, 2011
|
|
7LZ5
 
 | |
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
3C7F
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
8R8P
 
 | |
8R62
 
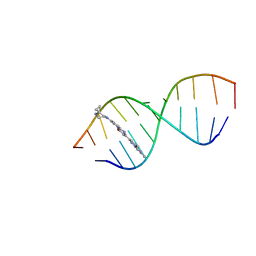 | | Solution structure of Risdiplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 7-(4,7-diazaspiro[2.5]octan-7-yl)-2-(2,8-dimethylimidazo[1,2-b]pyridazin-6-yl)-1~{H}-pyrido[1,2-a]pyrimidin-4-one, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8R63
 
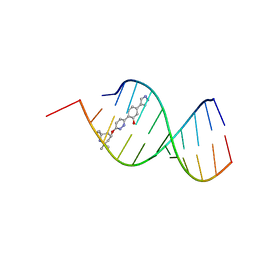 | | Solution structure of branaplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 5-(1~{H}-pyrazol-4-yl)-2-[6-(2,2,6,6-tetramethylpiperidin-4-yl)oxypyridazin-3-yl]phenol, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8CF2
 
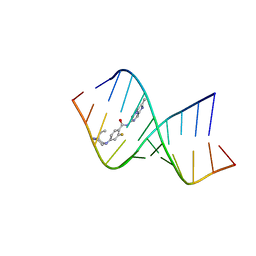 | | Solution structure of the RNA helix formed by the 5'-end of U1 snRNA and an A-1 bulged 5'-splice site in complex with SMN-CY | | Descriptor: | 4-[(3~{S})-3-ethylpiperazin-1-yl]-2-fluoranyl-~{N}-(2-methylimidazo[1,2-a]pyrazin-6-yl)benzamide, RNA (5'-R(P*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Marquevielle, J, Campagne, S. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
3JBJ
 
 | | Cryo-EM reconstruction of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBK
 
 | | Cryo-EM reconstruction of the metavinculin-actin interface | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBI
 
 | | MDFF model of the vinculin tail domain bound to F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
7MLK
 
 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
6VO7
 
 | |
9FTW
 
 | |
7P08
 
 | |
2LAU
 
 | |
6EZ6
 
 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
6EYZ
 
 | | PI3 kinase delta in complex with 4-Fluorophenyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylic acid, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
1TJY
 
 | | Crystal Structure of Salmonella typhimurium AI-2 receptor LsrB in complex with R-THMF | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, sugar transport protein | | Authors: | Miller, S.T, Xavier, K.B, Campagna, S.R, Taga, M.E, Semmelhack, M.F, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2
Mol.Cell, 15, 2004
|
|
1TM2
 
 | | Crystal Structure of the apo form of the Salmonella typhimurium AI-2 receptor LsrB | | Descriptor: | sugar transport protein | | Authors: | Miller, S.T, Xavier, K.B, Campagna, S.R, Taga, M.E, Semmelhack, M.F, Bassler, B.L, Hughson, F.M. | | Deposit date: | 2004-06-10 | | Release date: | 2004-09-28 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Salmonella typhimurium Recognizes a Chemically Distinct Form of the Bacterial Quorum-Sensing Signal AI-2
Mol.Cell, 15, 2004
|
|
1KZ7
 
 | | Crystal Structure of the DH/PH Fragment of Murine Dbs in Complex with the Placental Isoform of Human Cdc42 | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
1KZG
 
 | | DbsCdc42(Y889F) | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
