7KJ2
 
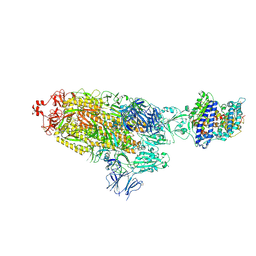 | | SARS-CoV-2 Spike Glycoprotein with one ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KRR
 
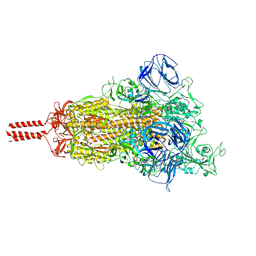 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRS
 
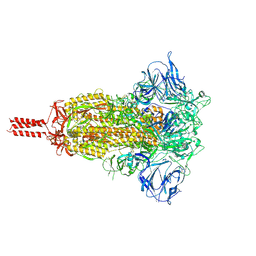 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRQ
 
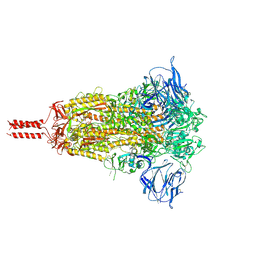 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
8AZS
 
 | | Type I amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
1R7T
 
 | | Glycosyltransferase A in complex with 3-deoxy-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7X
 
 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-amino-3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7Y
 
 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7U
 
 | | Glycosyltransferase B in complex with 3-deoxy-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R82
 
 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor, and uridine diphosphate-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
4NSQ
 
 | | Crystal structure of PCAF | | Descriptor: | COENZYME A, Histone acetyltransferase KAT2B | | Authors: | Lin, J.Y, Cai, Y.F. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3108 Å) | | Cite: | Dimeric structure of p300/CBP associated factor.
Bmc Struct.Biol., 14, 2014
|
|
3CPM
 
 | | plant peptide deformylase PDF1B crystal structure | | Descriptor: | Peptide deformylase, chloroplast, SULFATE ION, ... | | Authors: | Rodgers, D.W, Houtz, R.L, Dirk, L.M.A, Schmidt, J.J, Cai, Y. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture
Biochem.J., 413, 2008
|
|
4Q1W
 
 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, ASPARTYL PROTEASE, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
4Q1Y
 
 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
4Q1X
 
 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ASPARTYL PROTEASE, GLYCEROL, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
2MIW
 
 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MIV
 
 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2LZK
 
 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
