4DMM
 
 | |
4IKA
 
 | | Crystal structure of EV71 3Dpol-VPg | | Descriptor: | 3Dpol, NICKEL (II) ION, VPg | | Authors: | Chen, C, Wang, Y.X, Lou, Z.Y. | | Deposit date: | 2012-12-25 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of enterovirus 71 RNA-dependent RNA polymerase complexed with its protein primer VPg: implication for a trans mechanism of VPg uridylylation
J.Virol., 87, 2013
|
|
4OJ5
 
 | |
4DML
 
 | |
3US0
 
 | |
2MTE
 
 | | Solution structure of Doc48S | | Descriptor: | CALCIUM ION, Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation.
J.Struct.Biol., 188, 2014
|
|
4OJO
 
 | |
4OJ6
 
 | |
4OJP
 
 | |
4OJL
 
 | |
3US1
 
 | |
4KU4
 
 | |
8SOE
 
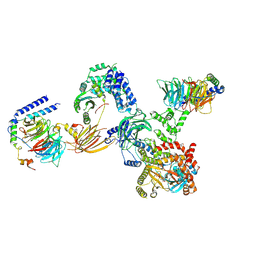 | |
8SOD
 
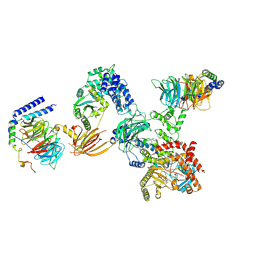 | |
8K32
 
 | |
8SO9
 
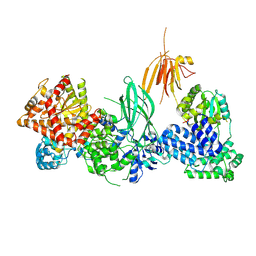 | | Phosphoinositide phosphate 3 kinase gamma | | Descriptor: | Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SOA
 
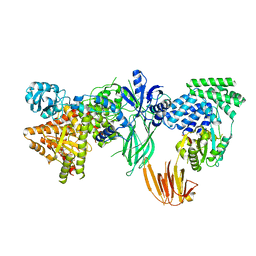 | | Phosphoinositide phosphate 3 kinase gamma bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SOC
 
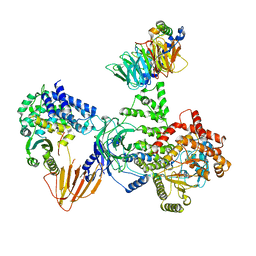 | | Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SOB
 
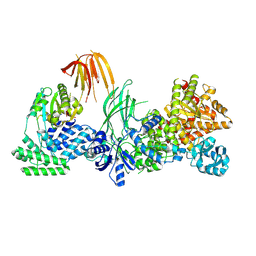 | | Phosphoinositide phosphate 3 kinase gamma bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3HMY
 
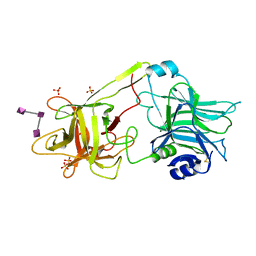 | | Crystal structure of HCR/T complexed with GT2 | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, SULFATE ION, ... | | Authors: | Chen, C, Fu, Z, Kim, J.-J.P, Barbieri, J.T, Baldwin, M.R. | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gangliosides as high affinity receptors for tetanus neurotoxin.
J.Biol.Chem., 284, 2009
|
|
3HN1
 
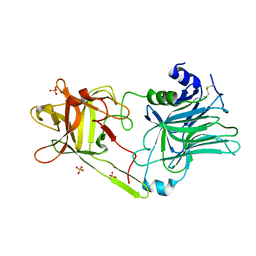 | | Crystal structure of HCR/T complexed with GT2 and lactose | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, SULFATE ION, Tetanus toxin, ... | | Authors: | Chen, C, Fu, Z, Kim, J.-J.P, Barbieri, J.T, Baldwin, M.R. | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gangliosides as high affinity receptors for tetanus neurotoxin.
J.Biol.Chem., 284, 2009
|
|
3ORF
 
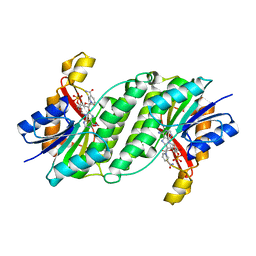 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
6JD1
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADH, and CPD at pH7.5 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JCZ
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADPH, and CPD at pH7.5 | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
3QYN
 
 | |
