7CX1
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine | | 分子名称: | 4-[(2R)-2-(methylamino)propyl]phenol, Decarboxylase | | 著者 | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | 登録日 | 2020-09-01 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine
to be published
|
|
7CWX
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis | | 分子名称: | DI(HYDROXYETHYL)ETHER, Decarboxylase, GLYCEROL | | 著者 | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | 登録日 | 2020-09-01 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis
to be published
|
|
7E58
 
 | | interferon-inducible anti-viral protein 2 | | 分子名称: | Guanylate-binding protein 2 | | 著者 | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5Z7J
 
 | | Crystal structure of a lactonase double mutant in complex with ligand l | | 分子名称: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, DI(HYDROXYETHYL)ETHER, Lactonase for protein | | 著者 | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-01-29 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
4NYL
 
 | |
8J3P
 
 | | Formate dehydrogenase mutant from from Candida dubliniensis M4 complexed with NADP+ | | 分子名称: | Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ma, W, Zheng, Y.C, Geng, Q, Chen, C, Xu, J.H. | | 登録日 | 2023-04-17 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
5XO7
 
 | | Crystal structure of a novel ZEN lactonase mutant with ligand a | | 分子名称: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, lactonase for protein | | 著者 | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2017-05-27 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
2JOE
 
 | | NMR Structure of E. Coli YehR Protein. Northeast Structural Genomics Target ER538. | | 分子名称: | Hypothetical lipoprotein yehR | | 著者 | Ding, K, Ramelot, T.A, Cort, J.R, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-03-08 | | 公開日 | 2007-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of E. Coli YehR Protein
To be Published
|
|
5XO8
 
 | | Crystal structure of a novel ZEN lactonase mutant with ligand Z | | 分子名称: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein | | 著者 | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2017-05-27 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5Z97
 
 | | Crystal structure of a lactonase double mutant in complex with ligand N | | 分子名称: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein, SULFATE ION | | 著者 | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-02-02 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5Z5J
 
 | | Crystal structure of a lactonase double mutant | | 分子名称: | DI(HYDROXYETHYL)ETHER, Lactonase for protein | | 著者 | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-01-18 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | 分子名称: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-26 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | 分子名称: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W1W
 
 | | NADPH-bound AKR4C17 mutant F291D | | 分子名称: | AKR4-2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | 登録日 | 2021-11-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7W1X
 
 | | Crystal structure of AKR4C16 bound with NADPH | | 分子名称: | AKR4-1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | 登録日 | 2021-11-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
2K5K
 
 | | Solution structure of RhR2 from Rhodobacter Sphaeroides. Northeast Structural Genomics Consortium | | 分子名称: | uncharacterized protein RhR2 | | 著者 | Lee, H, Bansal, S, Chen, C.X, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-28 | | 公開日 | 2008-08-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of RhR2
To be Published
|
|
7Y9J
 
 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium in complex with 5-nitro-1,2-benzisoxazole | | 分子名称: | 5-nitro-1,2-benzoxazole, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-24 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
7Y9K
 
 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium | | 分子名称: | Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-25 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
6LXM
 
 | | Crystal structure of C-terminal DNA-binding domain of Escherichia coli OmpR as a domain-swapped dimer | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Sadotra, S, Chen, C, Hsu, C.H. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.412 Å) | | 主引用文献 | Structural basis for promoter DNA recognition by the response regulator OmpR.
J.Struct.Biol., 213, 2020
|
|
5YVP
 
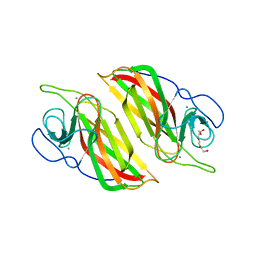 | | Crystal structure of an apo form cyclase Filc1 from Fischerella sp. TAU | | 分子名称: | CALCIUM ION, TETRAETHYLENE GLYCOL, cyclase A | | 著者 | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2017-11-27 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.051 Å) | | 主引用文献 | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z53
 
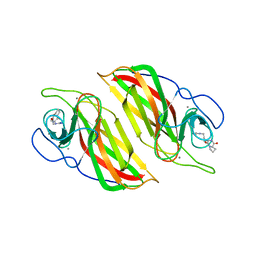 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with cyclo-L-Arg-D-Pro | | 分子名称: | 12-epi-hapalindole U synthase, CALCIUM ION, SULFATE ION, ... | | 著者 | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-01-16 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z54
 
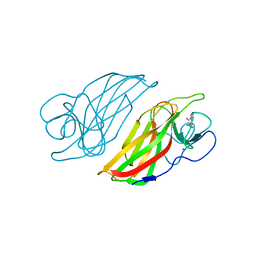 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 in complex with cyclo-L-Arg-D-Pro | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | 著者 | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2018-01-17 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2JRT
 
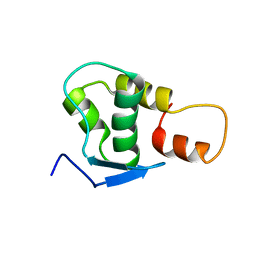 | | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides. Northeast Structural Genomics target RhR5 | | 分子名称: | Uncharacterized protein | | 著者 | Wang, L, Chen, C, Nwosu, C, Cunningham, K, Owens, L, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-28 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides.
To be Published
|
|
5YVL
 
 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, cyclase | | 著者 | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2017-11-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.059 Å) | | 主引用文献 | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | 分子名称: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | 登録日 | 2017-12-21 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.913 Å) | | 主引用文献 | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
