5WX9
 
 | | Crystal Structure of AtERF96 with GCC-box | | Descriptor: | Ethylene-responsive transcription factor ERF096, GCC-box motif | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2017-01-06 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box.
Plant Mol.Biol., 104, 2020
|
|
1KGF
 
 | |
1KGE
 
 | |
8SOE
 
 | |
8SOD
 
 | |
1GHM
 
 | | Structures of the acyl-enzyme complex of the staphylococcus aureus beta-lactamase mutant GLU166ASP:ASN170GLN with degraded cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, BETA-LACTAMASE, CARBONATE ION, ... | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2000-12-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of the acyl-enzyme complexes of the Staphylococcus aureus beta-lactamase mutant Glu166Asp:Asn170Gln with benzylpenicillin and cephaloridine.
Biochemistry, 40, 2001
|
|
8SOA
 
 | | Phosphoinositide phosphate 3 kinase gamma bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SOB
 
 | | Phosphoinositide phosphate 3 kinase gamma bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SO9
 
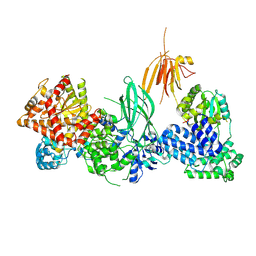 | | Phosphoinositide phosphate 3 kinase gamma | | Descriptor: | Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SOC
 
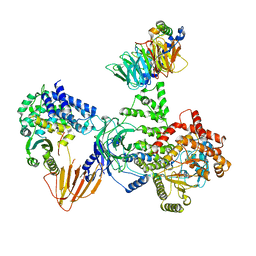 | | Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1BLH
 
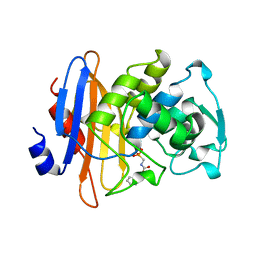 | |
1BLC
 
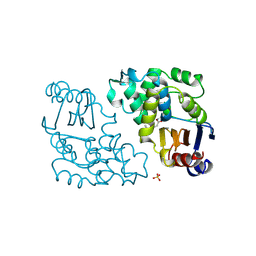 | |
1GHI
 
 | | STRUCTURE OF BETA-LACTAMASE GLU166ASP:ASN170GLN MUTANT | | Descriptor: | BETA-LACTAMASE, CARBONATE ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2000-12-18 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the acyl-enzyme complexes of the Staphylococcus aureus beta-lactamase mutant Glu166Asp:Asn170Gln with benzylpenicillin and cephaloridine.
Biochemistry, 40, 2001
|
|
1DJC
 
 | |
1NB2
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Bacillus Halodenitrificans | | Descriptor: | Nucleoside Diphosphate Kinase | | Authors: | Chen, C.-J, Liu, M.-Y, Chang, W.-C, Chang, T, Wang, B.-C, Le Gall, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nucleoside diphosphate kinase from Bacillus halodenitrificans: coexpression of its activity with a Mn-superoxide dismutase.
J.Struct.Biol., 142, 2003
|
|
1GHP
 
 | |
1KGG
 
 | | STRUCTURE OF BETA-LACTAMASE GLU166GLN:ASN170ASP MUTANT | | Descriptor: | PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relocation of the catalytic carboxylate group in class A beta-lactamase: the structure and function of the mutant enzyme Glu166-->Gln:Asn170-->Asp.
Protein Eng., 12, 1999
|
|
2X2N
 
 | | X-ray structure of cyp51 from trypanosoma brucei in complex with posaconazole in two different conformations | | Descriptor: | LANOSTEROL 14-ALPHA-DEMETHYLASE, POSACONAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, C.-K, Leung, S.S.F, Guilbert, C, Jacobson, M, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2010-01-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Cyp51 from Trypanosoma Cruzi and Trypanosoma Brucei Bound to the Antifungal Drugs Posaconazole and Fluconazole
Plos Negl Trop Dis, 4, 2010
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7N69
 
 | |
7N6A
 
 | |
7SJ4
 
 | | Human Trio residues 1284-1959 in complex with Rac1 | | Descriptor: | Ras-related C3 botulinum toxin substrate 1, Triple functional domain protein | | Authors: | Chen, C.-L, Ravala, S.K, Bandekar, S.J, Cash, J, Tesmer, J.J.G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural/functional studies of Trio provide insights into its configuration and show that conserved linker elements enhance its activity for Rac1.
J.Biol.Chem., 298, 2022
|
|
2ZYS
 
 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYI
 
 | | A. Fulgidus lipase with fatty acid fragment and calcium | | Descriptor: | CALCIUM ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYH
 
 | | mutant A. Fulgidus lipase S136A complexed with fatty acid fragment | | Descriptor: | CALCIUM ION, HEXADECANE, Lipase, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
