6L9H
 
 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6LX0
 
 | |
6LVP
 
 | |
6UGJ
 
 | |
6UGG
 
 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
6UGI
 
 | |
6URF
 
 | | Malic enzyme from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, NAD-dependent malic enzyme | | Authors: | Cuthbert, B.J, Burley, K.H, Goulding, C.W, Mathews, E.I, Beste, D.J. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Molecular Dynamics of Mycobacterium tuberculosis Malic Enzyme, a Potential Anti-TB Drug Target.
Acs Infect Dis., 7, 2021
|
|
6VEK
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length | | Descriptor: | contact-dependent immunity protein CdiI, contact-dependent toxin CdiA | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length
To Be Published
|
|
8PX2
 
 | |
8PXM
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH (1R,1'R)-7,7'-(pentane-1,5-diylbis(oxy))bis(1,3-dimethyl-1,3-dihydro-2H-benzo[d]azepin-2-one) | | Descriptor: | (1R)-7-[5-[[(1R)-1,3-dimethyl-2-oxidanylidene-1H-3-benzazepin-7-yl]oxy]pentoxy]-1,3-dimethyl-1H-3-benzazepin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Chung, C.W. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Design and Characterization of 1,3-Dihydro-2 H -benzo[ d ]azepin-2-ones as Rule-of-5 Compliant Bivalent BET Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8PX8
 
 | |
6VMY
 
 | | Structure of the B. subtilis cobalamin riboswitch | | Descriptor: | Adenosylcobalamin, B. subtilis cobalamin riboswitch, COBALT HEXAMMINE(III), ... | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an atypical cobalamin riboswitch reveals RNA structural adaptability as basis for promiscuous ligand binding.
Nucleic Acids Res., 48, 2020
|
|
8PVQ
 
 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
3UPU
 
 | | Crystal structure of the T4 Phage SF1B Helicase Dda | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3', ATP-dependent DNA helicase dda | | Authors: | He, X, Yun, M.K, Pemble IV, C.W, Kreuzer, K.N, Raney, K.D, White, S.W. | | Deposit date: | 2011-11-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation.
Structure, 20, 2012
|
|
8QHE
 
 | | Crystal structure IR-09 | | Descriptor: | 6-phosphogluconate dehydrogenase NADP-binding domain-containing protein, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biocatalysis in Drug Design: Engineered Reductive Aminases (RedAms) Are Used to Access Chiral Building Blocks with Multiple Stereocenters.
J.Am.Chem.Soc., 145, 2023
|
|
3UNO
 
 | |
1E95
 
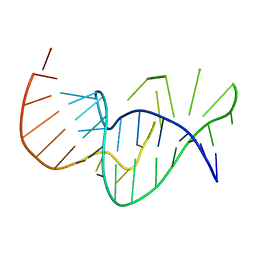 | | Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting | | Descriptor: | RNA (5'-(*GP*CP*GP*GP*CP*CP*AP*GP*CP*UP*CP* CP*AP*GP*GP*CP*CP*GP*CP*CP*AP*AP*AP*CP* AP*AP*UP*AP*UP*GP*GP*AP*GP*CP*AP*C)-3') | | Authors: | Michiels, P.J.A, Versleyen, A, Pleij, C.W.A, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2000-10-09 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pseudoknot of Srv-1 RNA, Involved in Ribosomal Frameshifting
J.Mol.Biol., 310, 2001
|
|
7AEA
 
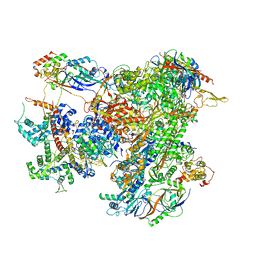 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8HGJ
 
 | |
7AE1
 
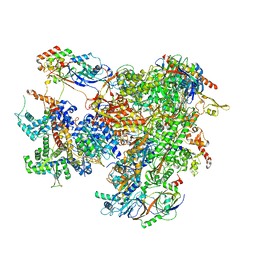 | | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8I9M
 
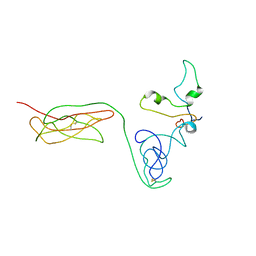 | | The RAGE and HMGB1 complex | | Descriptor: | Advanced glycosylation end product-specific receptor, High mobility group protein B1 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.19 Å) | | Cite: | The RAGE and HMGB1 complex
To Be Published
|
|
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8I9J
 
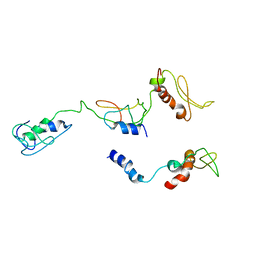 | | The PKR and E3L complex | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase, RNA-binding protein E3 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.39 Å) | | Cite: | Structural study of novel vaccinia virus E3L and dsRNA-dependent protein kinase complex.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
7A6H
 
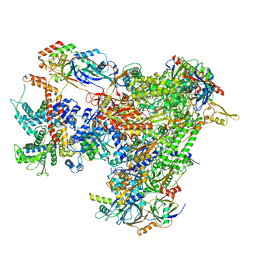 | | Cryo-EM structure of human apo RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-08-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8G6H
 
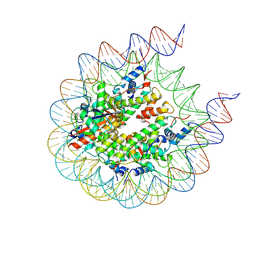 | | H2BK120ub+H3K79me2-modified nucleosome ubiquitin position 6 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
