5FTA
 
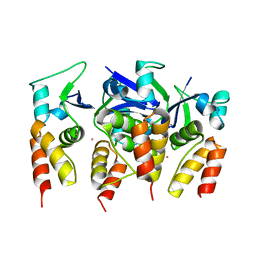 | | Crystal structure of the N-terminal BTB domain of human KCTD10 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 3, MERCURY (II) ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Tallant, C, Newman, J.A, Kopec, J, Fitzpatrick, F, Talon, R, Collins, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
2I0H
 
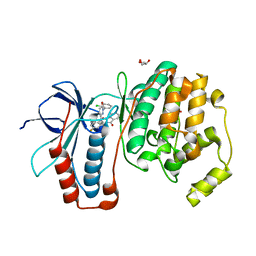 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5F92
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FFX
 
 | |
5FJQ
 
 | | Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus | | Descriptor: | CARBOHYDRATE BINDING PROTEIN, PUTATIVE, CPB33A, ... | | Authors: | Forsberg, Z, Nelson, C.E, Dalhus, B, Mekasha, S, Loose, J.S.M, Rohr, A.K, Eijsink, V.G.H, Gardner, J.G, Vaaje-Kolstad, G. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio Japonicus
J.Biol.Chem., 291, 2016
|
|
2H7V
 
 | | Co-crystal structure of YpkA-Rac1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Migration-inducing protein 5, ... | | Authors: | Prehna, G, Ivanov, M, Bliska, J.B, Stebbins, C.E. | | Deposit date: | 2006-06-04 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Yersinia virulence depends on mimicry of host rho-family nucleotide dissociation inhibitors.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HFN
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | FLAVIN MONONUCLEOTIDE, Synechocystis Photoreceptor (Slr1694) | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
5FFZ
 
 | |
4Y95
 
 | | Crystal structure of the kinase domain of Bruton's tyrosine kinase with mutations in the activation loop | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, BETA-MERCAPTOETHANOL, ... | | Authors: | Wang, Q, Rosen, C.E, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
2H7O
 
 | | Crystal structure of the Rho-GTPase binding domain of YpkA | | Descriptor: | Protein kinase ypkA | | Authors: | Prehna, G, Ivanov, M, Bliska, J.B, Stebbins, C.E. | | Deposit date: | 2006-06-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yersinia virulence depends on mimicry of host rho-family nucleotide dissociation inhibitors.
Cell(Cambridge,Mass.), 126, 2006
|
|
5J8C
 
 | | Human MOF C316S, E350Q crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, SODIUM ION, ... | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
5J8F
 
 | | Human MOF K274P crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, ZINC ION | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
2GWM
 
 | |
5G3Q
 
 | | Crystal structure of a hypothetical domain in WNK1 | | Descriptor: | WNK1 | | Authors: | Pinkas, D.M, Bufton, J.C, Sanvitale, C.E, Bartual, S.G, Adamson, R.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Crystal Structure of a Hypothetical Domain in Wnk1
To be Published
|
|
5E6Q
 
 | | Importin alpha binding to XRCC1 NLS peptide | | Descriptor: | CHLORIDE ION, DNA repair protein XRCC1 NLS peptide, GLYCEROL, ... | | Authors: | Pedersen, L.C, Kirby, T.W, Gassman, N.R, Smith, C.E, Gabel, S.A, Sobhany, M, Wilson, S.H, London, R.E. | | Deposit date: | 2015-10-10 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Nuclear Localization of the DNA Repair Scaffold XRCC1: Uncovering the Functional Role of a Bipartite NLS.
Sci Rep, 5, 2015
|
|
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YX5
 
 | | SpaO(SPOA1,2) | | Descriptor: | CHLORIDE ION, Surface presentation of antigens protein SpaO | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9001 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
4YFK
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFN
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
5FB2
 
 | |
4YX1
 
 | | SpaO(SPOA2) | | Descriptor: | CALCIUM ION, Surface presentation of antigens protein SpaO | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
4YX7
 
 | |
4YXC
 
 | |
