2AX6
 
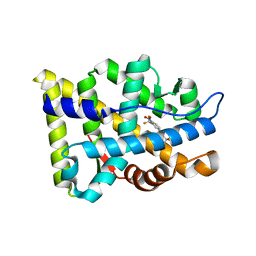 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain T877A Mutant In Complex With Hydroxyflutamide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2AXA
 
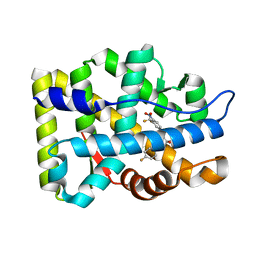 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain In Complex With S-1 | | Descriptor: | Androgen receptor, S-3-(4-FLUOROPHENOXY)-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
4HEH
 
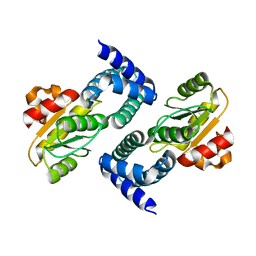 | |
1Z95
 
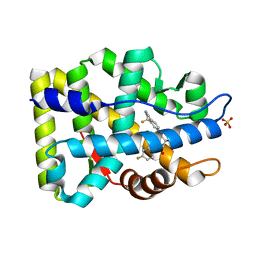 | | Crystal Structure of the Androgen Receptor Ligand-binding Domain W741L Mutant Complex with R-bicalutamide | | Descriptor: | Androgen Receptor, R-BICALUTAMIDE, SULFATE ION | | Authors: | Bohl, C.E, Gao, W, Miller, D.D, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for antagonism and resistance of bicalutamide in prostate cancer.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1HK5
 
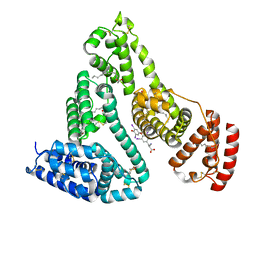 | | HUMAN SERUM ALBUMIN MUTANT R218H COMPLEXED WITH THYROXINE (3,3',5,5'-TETRAIODO-L-THYRONINE) and myristic acid (tetradecanoic acid) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HK4
 
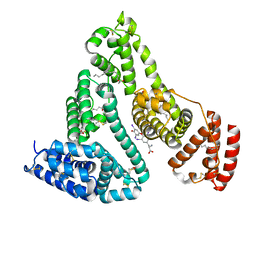 | | HUMAN SERUM ALBUMIN COMPLEXED WITH THYROXINE (3,3',5,5'-TETRAIODO-L-THYRONINE) and myristic acid (tetradecanoic acid) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HK1
 
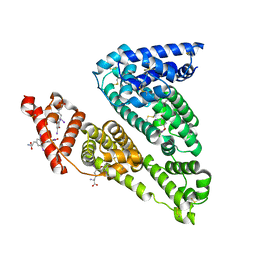 | | HUMAN SERUM ALBUMIN COMPLEXED WITH THYROXINE (3,3',5,5'-TETRAIODO-L-THYRONINE) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HK3
 
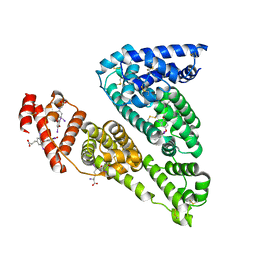 | | Human serum albumin mutant r218p complexed with thyroxine (3,3',5,5'-tetraiodo-l-thyronine) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1HK2
 
 | | HUMAN SERUM ALBUMIN MUTANT R218H COMPLEXED WITH THYROXINE (3,3',5,5'-TETRAIODO-L-THYRONINE) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SERUM ALBUMIN | | Authors: | Petitpas, I, Petersen, C.E, Ha, C.E, Bhattacharya, A.A, Zunszain, P.A, Ghuman, J, Bhagavan, N.V, Curry, S. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Albumin-Thyroxine Interactions and Familial Dysalbuminemic Hyperthyroxinemia
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2OZ7
 
 | | Crystal structure of the human androgen receptor T877A mutant ligand-binding domain with cyproterone acetate | | Descriptor: | Androgen receptor, CYPROTERONE ACETATE | | Authors: | Bohl, C.E, Wu, Z, Miller, D.D, Bell, C.E, Dalton, J.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the T877A human androgen receptor ligand-binding domain complexed to cyproterone acetate provides insight for ligand-induced conformational changes and structure-based drug design.
J.Biol.Chem., 282, 2007
|
|
4W8P
 
 | | Crystal structure of RIAM TBS1 in complex with talin R7R8 domains | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Chang, Y.C.E, Zhang, H, Wu, J. | | Deposit date: | 2014-08-25 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Insights into the Recruitment of Talin by RIAM in Integrin Signaling.
Structure, 22, 2014
|
|
6IEJ
 
 | | The C2 domain of cytosolic phospholipase A2 alpha bound to phosphatidylcholine | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CALCIUM ION, Cytosolic phospholipase A2, ... | | Authors: | Hirano, Y, Gao, Y.G, Stephenson, D.J, Vu, N.T, Malinina, L, Chalfant, C.E, Patel, D.J, Brown, R.E. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2alpha.
Elife, 8, 2019
|
|
8QDC
 
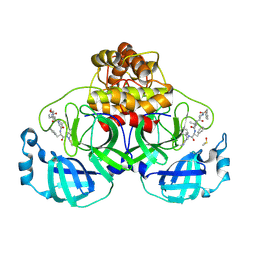 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3642 (compound 1 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(2~{S})-1-[[iminomethyl-(phenylmethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of an active-site titrant for SARS-CoV-2 main protease as an indispensable tool for evaluating enzyme kinetics.
Acta Pharm Sin B, 14, 2024
|
|
2AX9
 
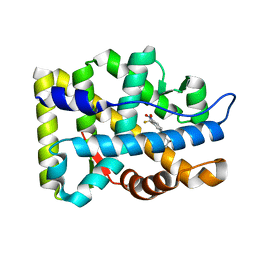 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain In Complex With R-3 | | Descriptor: | (R)-3-BROMO-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE, Androgen receptor | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2AX7
 
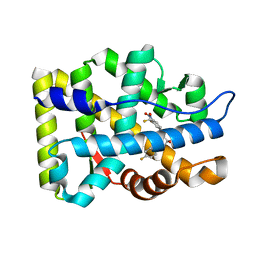 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain T877A Mutant In Complex With S-1 | | Descriptor: | Androgen receptor, S-3-(4-FLUOROPHENOXY)-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2AX8
 
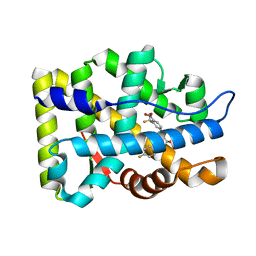 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain W741L Mutant In Complex With S-1 | | Descriptor: | Androgen receptor, S-3-(4-FLUOROPHENOXY)-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
6B72
 
 | | A novel HIV-1 Nef dimer interface induced by a single octyl-glucoside molecule | | Descriptor: | Protein Nef, octyl beta-D-glucopyranoside | | Authors: | Wu, M, Augelli-Szafran, C.E, Ptak, R.G, Smithgall, T.E. | | Deposit date: | 2017-10-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A single beta-octyl glucoside molecule induces HIV-1 Nef dimer formation in the absence of partner protein binding.
PLoS ONE, 13, 2018
|
|
3RLJ
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with SARM S-22 | | Descriptor: | (2S)-3-(4-cyanophenoxy)-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | Authors: | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
4LMF
 
 | | C1s CUB1-EGF-CUB2 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain, SODIUM ION | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.921 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3RBH
 
 | | Structure of alginate export protein AlgE from Pseudomonas aeruginosa | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, Alginate production protein AlgE, ... | | Authors: | Whitney, J.C, Hay, I.D, Li, C, Eckford, P.D, Robinson, H, Amaya, M.F, Wood, L.F, Ohman, D.E, Bear, C.E, Rehm, B.H, Howell, P.L. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-27 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for alginate secretion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KQ5
 
 | |
4M1P
 
 | |
6P1K
 
 | | Cryo-EM structure of Escherichia coli sigma70 bound RNAP polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6P2T
 
 | | BshB from Bacillus subtilis complexed with citrate | | Descriptor: | CITRIC ACID, N-acetyl-alpha-D-glucosaminyl L-malate deacetylase 1, SODIUM ION, ... | | Authors: | Cook, P.D, Meloche, C.E. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | X-ray crystallographic structure of BshB, the zinc-dependent deacetylase involved in bacillithiol biosynthesis.
Protein Sci., 29, 2020
|
|
