2H7V
 
 | | Co-crystal structure of YpkA-Rac1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Migration-inducing protein 5, ... | | Authors: | Prehna, G, Ivanov, M, Bliska, J.B, Stebbins, C.E. | | Deposit date: | 2006-06-04 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Yersinia virulence depends on mimicry of host rho-family nucleotide dissociation inhibitors.
Cell(Cambridge,Mass.), 126, 2006
|
|
6ELC
 
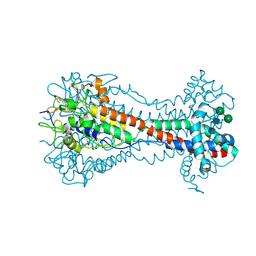 | | Crystal Structure of O-linked Glycosylated VSG3 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stebbins, C.E. | | Deposit date: | 2017-09-28 | | Release date: | 2018-07-11 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat.
Nat Microbiol, 3, 2018
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
1USL
 
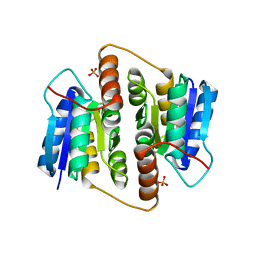 | | Structure Of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, Complexed With Phosphate. | | Descriptor: | PHOSPHATE ION, RIBOSE 5-PHOSPHATE ISOMERASE B | | Authors: | Roos, A.K, Andersson, C.E, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-11-25 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase Has a Known Fold, But a Novel Active Site
J.Mol.Biol., 335, 2004
|
|
1V1P
 
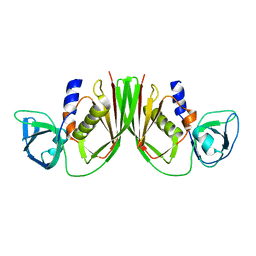 | |
1B8Z
 
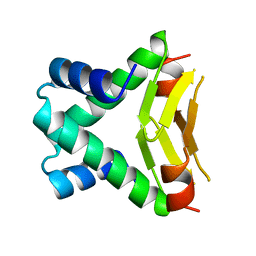 | | HU FROM THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (HISTONELIKE PROTEIN HU) | | Authors: | Christodoulou, E, Rypniewski, W.R, Vorgias, C.E. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, overproduction, purification and crystallization of the DNA binding protein HU from the hyperthermophilic eubacterium Thermotoga maritima.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6GRI
 
 | | E. coli Microcin synthetase McbBCD complex | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Microcin B17-processing protein McbB, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
1VCB
 
 | | THE VHL-ELONGINC-ELONGINB STRUCTURE | | Descriptor: | PROTEIN (ELONGIN B), PROTEIN (ELONGIN C), PROTEIN (VHL) | | Authors: | Stebbins, C.E, Kaelin, W.G, Pavletich, N.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function.
Science, 284, 1999
|
|
6GY5
 
 | | Crystal structure of the kelch domain of human KLHL20 in complex with DAPK1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Chen, Z, Hozjan, V, Strain-Damerell, C, Williams, E, Wang, D, Cooper, C.D.O, Sanvitale, C.E, Fairhead, M, Carpenter, E.P, Pike, A.C.W, Krojer, T, Srikannathasan, V, Sorrell, F, Johansson, C, Mathea, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Structural Basis for Recruitment of DAPK1 to the KLHL20 E3 Ligase.
Structure, 27, 2019
|
|
2HNY
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
1ATG
 
 | | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PERIPLASMIC MOLYBDATE-BINDING PROTEIN, ... | | Authors: | Lawson, D.M, Pau, R.N, Williams, C.E.M, Mitchenall, L.A. | | Deposit date: | 1997-08-14 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA.
Structure, 6, 1998
|
|
2HFO
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | Activator of photopigment and puc expression, FLAVIN MONONUCLEOTIDE | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HO0
 
 | |
1BTQ
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
6EIX
 
 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
1SZH
 
 | | Crystal Structure of C. elegans HER-1 | | Descriptor: | ACETATE ION, Her-1 protein | | Authors: | Hamaoka, B.Y, Dann III, C.E, Geisbrecht, B.V, Leahy, D.J. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6GQT
 
 | | KRAS-169 Q61H GPPNHP + PPIN-2 | | Descriptor: | CITRIC ACID, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6GRH
 
 | | E. coli Microcin synthetase McbBCD complex with truncated pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
1BWF
 
 | | ESCHERICHIA COLI GLYCEROL KINASE MUTANT WITH BOUND ATP ANALOG SHOWING SUBSTANTIAL DOMAIN MOTION | | Descriptor: | GLYCEROL, GLYCEROL KINASE, MAGNESIUM ION, ... | | Authors: | Bystrom, C.E, Pettigrew, D.W, Branchaud, B.P, Remington, S.J. | | Deposit date: | 1998-09-23 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Escherichia coli glycerol kinase variant S58-->W in complex with nonhydrolyzable ATP analogues reveal a putative active conformation of the enzyme as a result of domain motion.
Biochemistry, 38, 1999
|
|
1BTT
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTR
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1993-05-25 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
6GOD
 
 | | KRAS full length wild-type GPPNHP | | Descriptor: | GLYCEROL, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Ehebauer, M.T, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1CA1
 
 | |
