3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2A33
 
 | | X-Ray Structure of a Lysine Decarboxylase-Like Protein from Arabidopsis Thaliana Gene AT2G37210 | | Descriptor: | MAGNESIUM ION, SULFATE ION, hypothetical protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structures of the conserved hypothetical proteins from Arabidopsis thaliana gene loci At5g11950 and AT2g37210.
Proteins, 65, 2006
|
|
3RB8
 
 | | Structure of the phage tubulin PhuZ(SeMet)-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Agard, D.A, Pogliano, J, Kraemer, J.A, Erb, M.L, Waddling, C.A, Montabana, E.A, Wang, H, Nguyen, K, Pham, S. | | Deposit date: | 2011-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell.
Cell(Cambridge,Mass.), 149, 2012
|
|
2A18
 
 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
3TSI
 
 | | Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) stalk domain | | Descriptor: | Hemagglutinin-neuraminidase | | Authors: | Bose, S, Welch, B.D, Kors, C.A, Yuan, P, Jardetzky, T.S, Lamb, R.A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion.
J.Virol., 85, 2011
|
|
2A4H
 
 | | Solution structure of Sep15 from Drosophila melanogaster | | Descriptor: | Selenoprotein Sep15 | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
2A2P
 
 | | Solution structure of SelM from Mus musculus | | Descriptor: | Selenoprotein M | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
3U27
 
 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
3TYV
 
 | |
3U64
 
 | | The Crystal Structure of Tat-T (Tp0956) | | Descriptor: | Protein TP_0956, SULFATE ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
3UBY
 
 | | Crystal structure of human alklyadenine DNA glycosylase in a lower and higher-affinity complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDC)P*TP*TP*GP*CP*CP*T)-3'), DNA-3-methyladenine glycosylase | | Authors: | Setser, J.W, Lingaraju, G.M, Davis, C.A, Samson, L.D, Drennan, C.L. | | Deposit date: | 2011-10-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Searching for DNA lesions: structural evidence for lower- and higher-affinity DNA binding conformations of human alkyladenine DNA glycosylase.
Biochemistry, 51, 2012
|
|
8E5C
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L50F with Nirmatrelvir captured in two conformational states | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8E4W
 
 | |
2YB9
 
 | | Crystal Structure of Human Neutral Endopeptidase complexed with a heteroarylalanine diacid. | | Descriptor: | HETEROARYLALANINE 5-PHENYL OXAZOLE, NEPRILYSIN, ZINC ION | | Authors: | Glossop, M.S, Bazin, R.J, Dack, K.N, Done, S, Fox, D.N.A, MacDonald, G.A, Mills, M, Owen, D.R, Phillips, C, Reeves, K.A, Ringer, T.J, Strang, R.S, Watson, C.A.L. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-25 | | Last modified: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Evaluation of Heteroarylalanine Diacids as Potent and Selective Neutral Endopeptidase Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8F5A
 
 | |
2YOJ
 
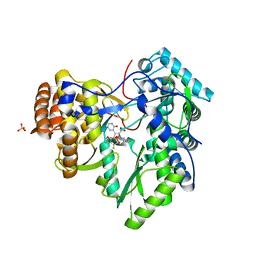 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
2LDO
 
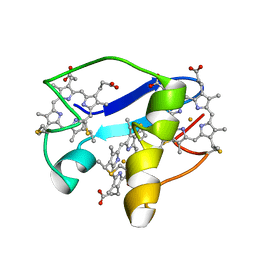 | | Solution structure of triheme cytochrome PpcA from Geobacter sulfurreducens reveals the structural origin of the redox-Bohr effect | | Descriptor: | Cytochrome c3, HEME C | | Authors: | Morgado, L, Paixao, V.B, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | Revealing the structural origin of the redox-Bohr effect: the first solution structure of a cytochrome from Geobacter sulfurreducens.
Biochem.J., 441, 2012
|
|
8ER6
 
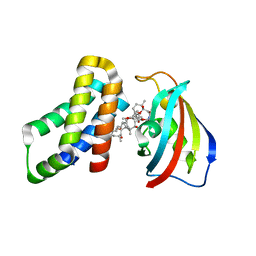 | | FKBP12-FRB in Complex with Compound 11 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1,2-ETHANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
1D79
 
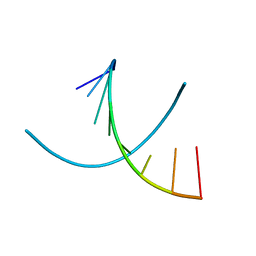 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
8ER7
 
 | | FKBP12-FRB in Complex with Compound 12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-9,27-dihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-5,10,21-trimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
1DBI
 
 | | CRYSTAL STRUCTURE OF A THERMOSTABLE SERINE PROTEASE | | Descriptor: | AK.1 SERINE PROTEASE, CALCIUM ION, SODIUM ION | | Authors: | Smith, C.A, Toogood, H.S, Baker, H.M, Daniel, R.M, Baker, E.N. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calcium-mediated thermostability in the subtilisin superfamily: the crystal structure of Bacillus Ak.1 protease at 1.8 A resolution.
J.Mol.Biol., 294, 1999
|
|
1DL8
 
 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
1DCG
 
 | | THE MOLECULAR STRUCTURE OF THE LEFT-HANDED Z-DNA DOUBLE HELIX AT 1.0 ANGSTROM ATOMIC RESOLUTION. GEOMETRY, CONFORMATION, AND IONIC INTERACTIONS OF D(CGCGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gessner, R.V, Frederick, C.A, Quigley, G.J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG).
J.Biol.Chem., 264, 1989
|
|
2LMZ
 
 | |
1EOK
 
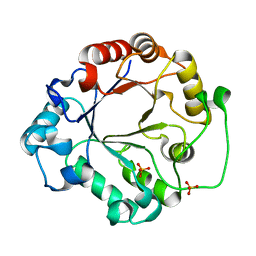 | | CRYSTAL STRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
