7UV9
 
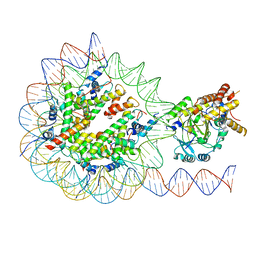 | | KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate | | Descriptor: | DNA (185-MER), FE (III) ION, Histone H2A type 1, ... | | Authors: | Spangler, C.J, Skrajna, A, Foley, C.A, Budziszewski, G.R, Azzam, D.N, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7UVA
 
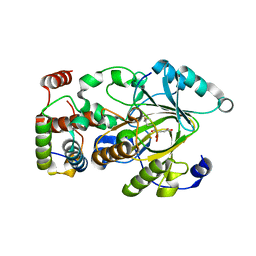 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
1MS6
 
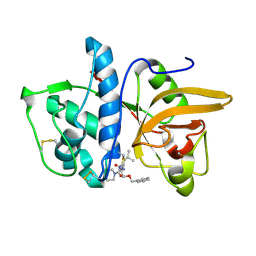 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | Descriptor: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | Authors: | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
4OPF
 
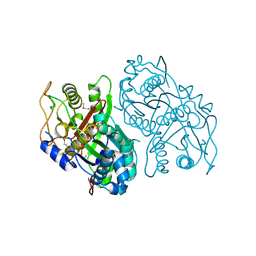 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1MY6
 
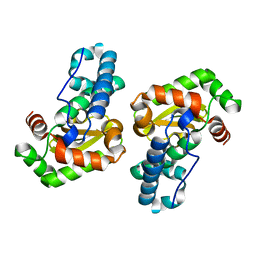 | | The 1.6 A Structure of Fe-Superoxide Dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus : Correlation of EPR and Structural Characteristics | | Descriptor: | FE (III) ION, Iron (III) Superoxide Dismutase | | Authors: | Yoshida, S.M, Cascio, D, Sawaya, M.R, Yeates, T.O, Kerfeld, C.A. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A resolution structure of Fe-superoxide dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus.
J.BIOL.INORG.CHEM., 8, 2003
|
|
3DCY
 
 | |
3DI2
 
 | |
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4OX7
 
 | |
1NP4
 
 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, PROTEIN (NITROPHORIN 4), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Weichsel, A, Champagne, D.E, Balfour, C.A, Montfort, W.R. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein.
Structure, 6, 1998
|
|
3E82
 
 | | Crystal structure of a putative oxidoreductase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Eswaramoorthy, S, Mohammad, M.B, Thomas, C.A, Brown, A.C, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-19 | | Release date: | 2008-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Klebsiella pneumoniae
To be Published
|
|
4OCW
 
 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
1NND
 
 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | Descriptor: | Reverse Transcriptase | | Authors: | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
6X5Y
 
 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
1NR7
 
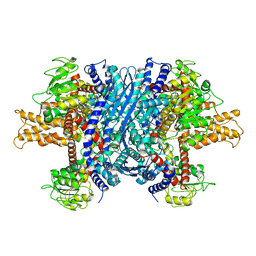 | | Crystal structure of apo bovine glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
3EA6
 
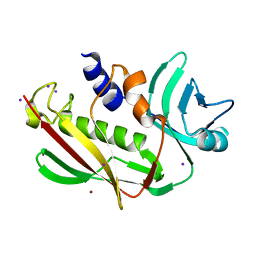 | | Atomic resolution of crystal structure of SEK | | Descriptor: | IODIDE ION, Staphylococcal enterotoxin K, ZINC ION | | Authors: | Shi, K, Huseby, M, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2008-08-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Studies of an Emerging Pyrogenic Superantigen, SEK
To be Published
|
|
4O89
 
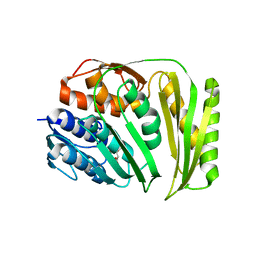 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | Descriptor: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
3EKX
 
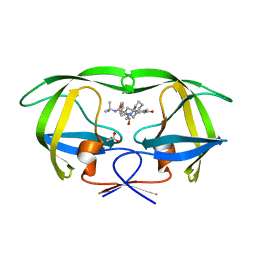 | |
6XAT
 
 | | Crystal structure of the human FoxP4 DNA binding Domain | | Descriptor: | FOXP4 protein, SODIUM ION | | Authors: | VIllalobos, P, Castro-Fernandez, V, Medina, E, Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unraveling the folding and dimerization properties of the human FoxP subfamily of transcription factors.
Febs Lett., 597, 2023
|
|
4ODH
 
 | | Crystal structure of human Fab CAP256-VRC26.UCA, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.UCA heavy chain, CAP256-VRC26.UCA light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
3EKQ
 
 | | Crystal structure of inhibitor saquinavir (SQV) in complex with multi-drug resistant HIV-1 protease (L63P/V82T/I84V) (referred to as ACT in paper) | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PHOSPHATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N.M, Schiffer, C.A, Nalivaika, E. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EM6
 
 | | Crystal structure of I50L/A71V mutant of hiv-1 protease in complex with inhibitor darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Royer, C.J, King, N.M, Prabu-Jeyabalan, M, Ng, C, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies on atazanavir specific I50L drug-resistant HIV-1 protease mutant.
To be Published
|
|
4DJO
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP56 | | Descriptor: | 2-[(dichloroacetyl)amino]ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, GLYCEROL, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
4DJQ
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP86 | | Descriptor: | 2-(2-oxoimidazolidin-1-yl)ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, PHOSPHATE ION, Pol polyprotein | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
1OR7
 
 | | Crystal Structure of Escherichia coli sigmaE with the Cytoplasmic Domain of its Anti-sigma RseA | | Descriptor: | RNA polymerase sigma-E factor, Sigma-E factor negative regulatory protein | | Authors: | Campbell, E.A, Tupy, J.L, Gruber, T.M, Wang, S, Sharp, M.M, Gross, C.A, Darst, S.A. | | Deposit date: | 2003-03-12 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA.
Mol.Cell, 11, 2003
|
|
